We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
SARS Coronavirus Main Proteinase
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
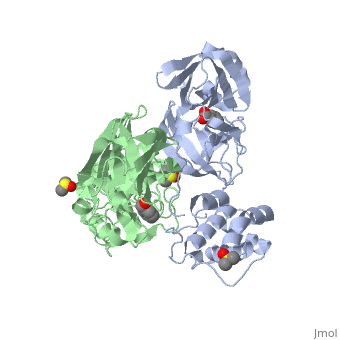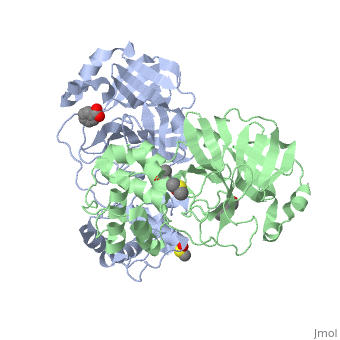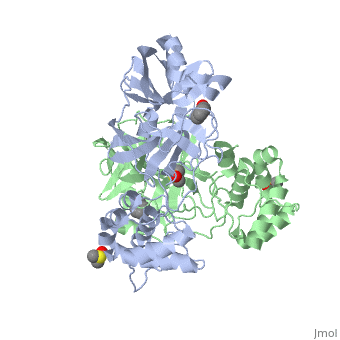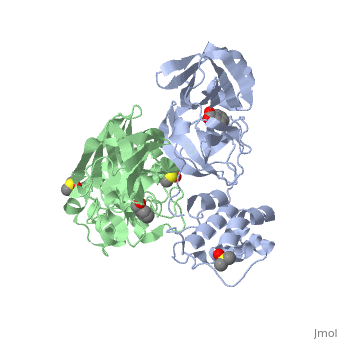
<StructureSection load='2vj1' size='350' side='right' caption='Human SARS coronavirus proteinase dimer complex with benzoic acid, dimethylamino benzoic acid and DMSO, [[2vj1]]' scene=''> | <StructureSection load='2vj1' size='350' side='right' caption='Human SARS coronavirus proteinase dimer complex with benzoic acid, dimethylamino benzoic acid and DMSO, [[2vj1]]' scene=''> | ||
==Introduction== | ==Introduction== | ||
| - | Before the emergence of the SARS-CoV, no research efforts were being directed toward the discovery of antiviral drugs against coronaviruses. The high infectivity and mortality rate established SARS as a significant global threat for which no efficacious therapy was available. Since the epidemic, which began in 2003, new insights into the genomics and pathogenisis of the SARS-CoV revealed several novel potential anti-coronavirus targets. Several proteins encoded by the SARS-CoV could be considered as targets for therapeutic intervention. The focus of this article is the '''SARS-CoV main protease'''. | + | Before the emergence of the SARS-CoV, no research efforts were being directed toward the discovery of antiviral drugs against coronaviruses. The high infectivity and mortality rate established SARS as a significant global threat for which no efficacious therapy was available. Since the epidemic, which began in 2003, new insights into the genomics and pathogenisis of the SARS-CoV revealed several novel potential anti-coronavirus targets. Several proteins encoded by the SARS-CoV could be considered as targets for therapeutic intervention. The focus of this article is the '''SARS-CoV main protease'''. Norovirus protease is an essential enzyme for proteoplytic maturation of norovirus nonstructural proteins and is implicated as a potential antiviral drug target<ref>PMID:23319456</ref>. |
==Coronaviruses== | ==Coronaviruses== | ||
Revision as of 18:18, 17 September 2018
| |||||||||||
3D structures of virus proteinase
Updated on 17-September-2018
References
- ↑ Takahashi D, Hiromasa Y, Kim Y, Anbanandam A, Yao X, Chang KO, Prakash O. Structural and dynamics characterization of norovirus protease. Protein Sci. 2013 Jan 15. doi: 10.1002/pro.2215. PMID:23319456 doi:http://dx.doi.org/10.1002/pro.2215
- ↑ Murray, Patrick R., Ken S. Rosenthal, and Michael A. Pfaller. "Coronaviruses and Noroviruses." Medical Microbiology. 6th ed. Philadelphia: Mosby/Elsevier, 2009. 565-68. Print.
- ↑ Ziebuhr, J., Herold,J., and Siddell, S.G. (1995). Characterization of a human coronavirus (strain 229E) 3C-like proteinase activity. J. Virol. 69, 4331-4338.
- ↑ Ksiazek TG, Erdman D, Goldsmith CS, et al. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med 2003;348:1953-1966
- ↑ "Summary of probable SARS cases with onset of illness from 1 November 2002 to 31 July 2003". WHO. Retrieved 2008-10-31.
- ↑ Li W, Shi Z, Yu M, et al. (2005). "Bats are natural reservoirs of SARS-like coronaviruses". Science (journal) 310 (5748): 676–9. doi:10.1126/science.1118391. PMID 16195424.
- ↑ Ziebuhr, J., Snijder, E.J., and Gorbaleya, A.E. (2000). Virus-encoded proteinases and proteolytic processing in Nidovirales. J. Gen. Virol. 81, 853-879.
- ↑ Anad, K., Ziebhr, J., Wadhani, P., Mesters, J.R., and Hilgenfeld, R. (2003). Coronavirus main protease (3CLPro) structure: basis for design of anti-SARS drugs. Science300, 1763-1767.
- ↑ Anad, K., Ziebhr, J., Wadhani, P., Mesters, J.R., and Hilgenfeld, R. (2003). Coronavirus main protease (3CLPro) structure: basis for design of anti-SARS drugs. Science300, 1763-1767.
- ↑ Verschueren, Koen H.G., Ksenia Pumpor, Stefan Anemüller, Shuai Chen, Jeroen R. Mesters, and Rolf Hilgenfeld. "A Structural View of the Inactivation of the SARS Coronavirus Main Proteinase by Benzotriazole Esters." Chemistry & Biology 15.6 (2008): 597-606. Web. Oct. 2010.
Proteopedia Page Contributors and Editors (what is this?)
Sarra Borhanian, Michal Harel, David Canner, Ann Taylor, Alexander Berchansky