We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1473
From Proteopedia
(Difference between revisions)
| Line 79: | Line 79: | ||
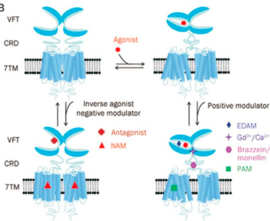
The kind and specificity of the agonists determine the activation pathway taken by GPCRs in terms of the energy ordering of the different states which is also premeditated by the respective activation energy landscapes. This means that the agonist-bound inactive state will have the lowest energy state among the various states within the degree of activation. | The kind and specificity of the agonists determine the activation pathway taken by GPCRs in terms of the energy ordering of the different states which is also premeditated by the respective activation energy landscapes. This means that the agonist-bound inactive state will have the lowest energy state among the various states within the degree of activation. | ||
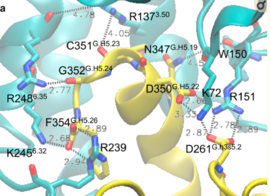
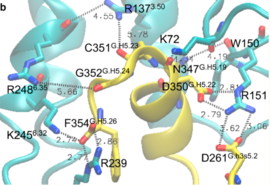
| - | Activation of the G-protein can be explained by interactions between the GPCR (specifically the ICD) and the G-alpha subunit. During analysis, Molecular Dynamics (MD) simulations found that there is formation of Salt-bridges and Hydrogen bonds between the C-terminal helix of G∝i and some regions in the ICD. Salt-bridges form between D261, D350 of G∝i and K72 of the IC loop 1 and R51 of IC loop 2. There's also formation of Hydrogen bonds between C351 and R137 of the G∝i that replace the inactive state R151-D136-R137-T247 network.Another salt-bridge is formed between the carboxylate group on the G∝i C-terminal residue F354 and K245 of the TM6 and R239 of the IC loop 3. All these structural changes facilitate ligand binding to receptor with high affinity and specificity and leads to an ordered state that further accommodates a more tighter binding to the ligand. The ICD then release the D/E R Y sequence motif from the TMD and this process is coupled to the binding of a ligand that is specific to the coupled domain (G-protein).Full activation occurs when G-proteins bind to the ICD. The stabilizing factor of the activated state of the GPCRs is attributed to the interactions between the Ga (G alpha subunit) and the GPCR. | + | Activation of the G-protein can be explained by interactions between the GPCR (specifically the ICD) and the G-alpha subunit. During analysis, Molecular Dynamics (MD) simulations found that there is formation of Salt-bridges and Hydrogen bonds between the C-terminal helix of G∝i and some regions in the ICD. Salt-bridges form between D261, D350 of G∝i and K72 of the IC loop 1 and R51 of IC loop 2.<ref>PMID: 27332120</ref> There's also formation of Hydrogen bonds between C351 and R137 of the G∝i that replace the inactive state R151-D136-R137-T247 network.Another salt-bridge is formed between the carboxylate group on the G∝i C-terminal residue F354 and K245 of the TM6 and R239 of the IC loop 3. All these structural changes facilitate ligand binding to receptor with high affinity and specificity and leads to an ordered state that further accommodates a more tighter binding to the ligand. The ICD then release the D/E R Y sequence motif from the TMD and this process is coupled to the binding of a ligand that is specific to the coupled domain (G-protein).Full activation occurs when G-proteins bind to the ICD. The stabilizing factor of the activated state of the GPCRs is attributed to the interactions between the Ga (G alpha subunit) and the GPCR. |
Revision as of 16:40, 10 December 2018
| This Sandbox is Reserved from November 5 2018 through January 1, 2019 for use in the course "CHEM 4923: Senior Project taught by Christina R. Bourne at the University of Oklahoma, Norman, USA. This reservation includes Sandbox Reserved 1471 through Sandbox Reserved 1478. |
To get started:
More help: Help:Editing |
This page is reserved for Manny
Your Heading Here (maybe something like 'Structure')
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
THE GENUIS OF GPCRs.
|
| |||||||||||