User:Gisele A. Andree/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 2: | Line 2: | ||
<StructureSection load='1stp' size='340' side='right' caption='Caption for this structure' scene=''> | <StructureSection load='1stp' size='340' side='right' caption='Caption for this structure' scene=''> | ||
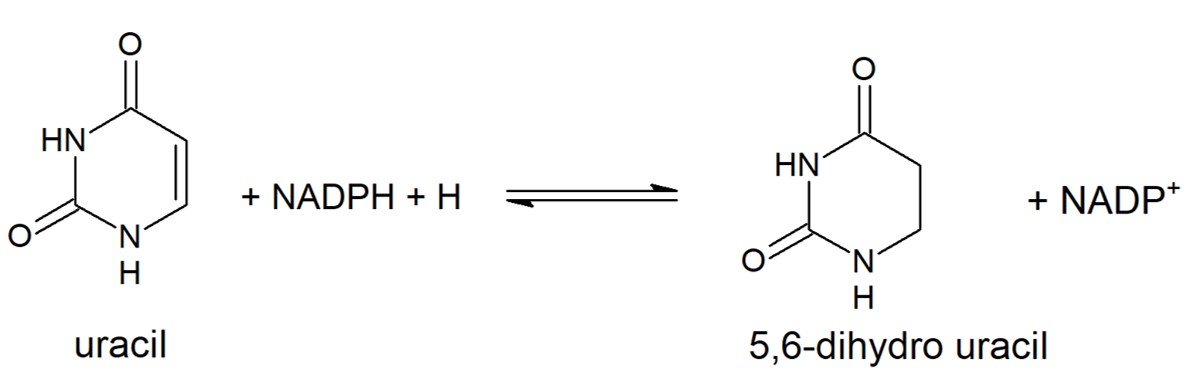
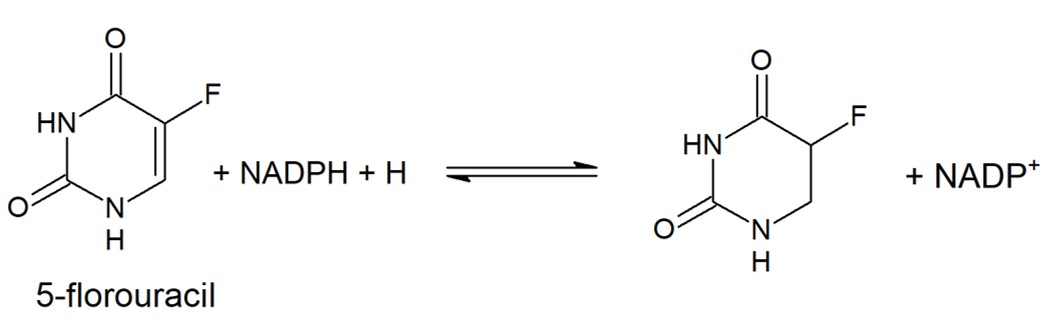
Dihydropyrimidine Dehydrogenase (DPD) is an important enzyme involved in the degradation of pyrimidines in the body. It is a 220 kDa protein that binds co-factors; FAD, FMN, [4Fe-4S] clusters, and substrates; NADPH and pyrimidines. | Dihydropyrimidine Dehydrogenase (DPD) is an important enzyme involved in the degradation of pyrimidines in the body. It is a 220 kDa protein that binds co-factors; FAD, FMN, [4Fe-4S] clusters, and substrates; NADPH and pyrimidines. | ||
| - | |||
| - | You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | ||
== Function == | == Function == | ||
| Line 35: | Line 33: | ||
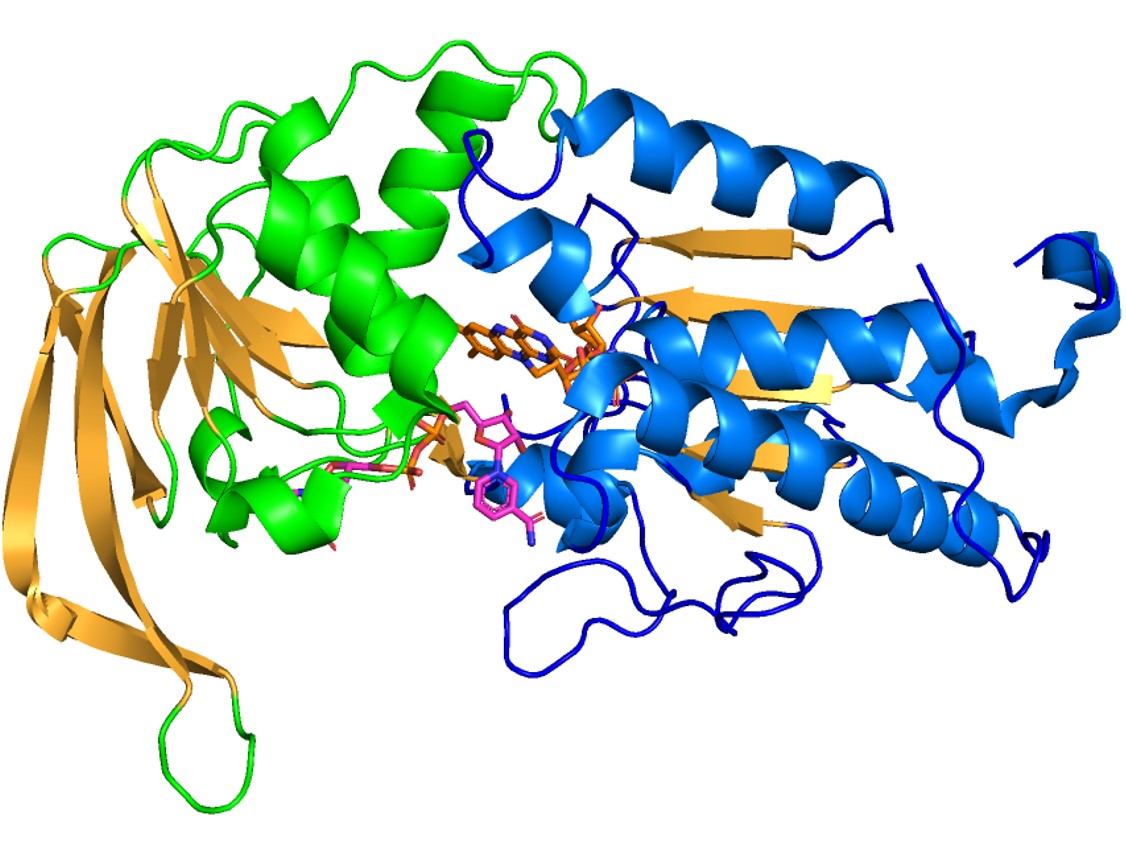
In the following figure, domain 2 is shown in blue, domain 3 is in green, FAD is colored in orange and NADPH is in pink. | In the following figure, domain 2 is shown in blue, domain 3 is in green, FAD is colored in orange and NADPH is in pink. | ||
| + | |||
| + | [[Image:D2_and_D3.jpg]] | ||
| + | |||
| + | '''Domain 4''' | ||
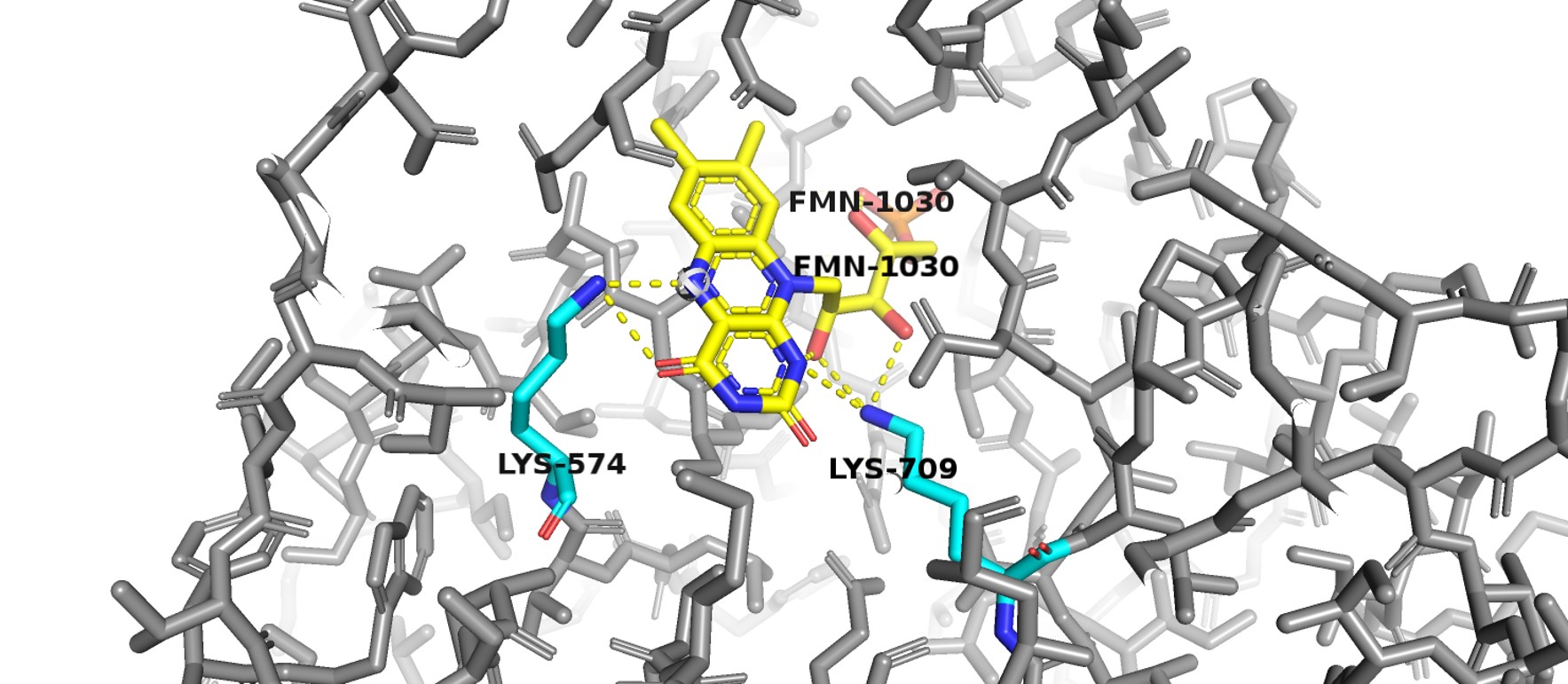
| + | Domain 4 has a typical α/β 8 barrel fold. It binds FMN towards the C-terminal end of the barrel forming strands. Most of the residues that have their side chains interacting with FMN are conserved between homologous structures. Two lysine residues (K574 and K709) bind to the isoalloxazine ring. Positively charged lysine 574 interacts with the redox active N5 atom of FMN ring. | ||
| + | |||
| + | [[Image:D4.jpg]] | ||
| + | |||
| + | '''Domain 5''' | ||
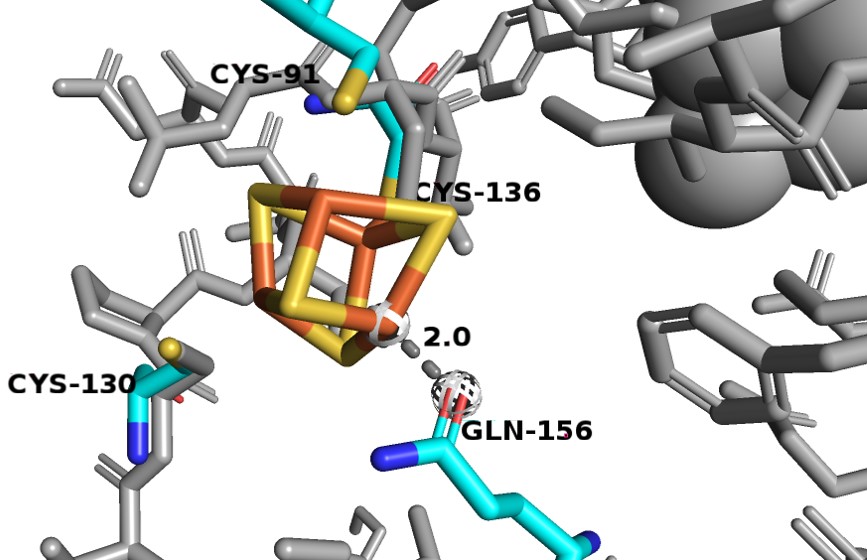
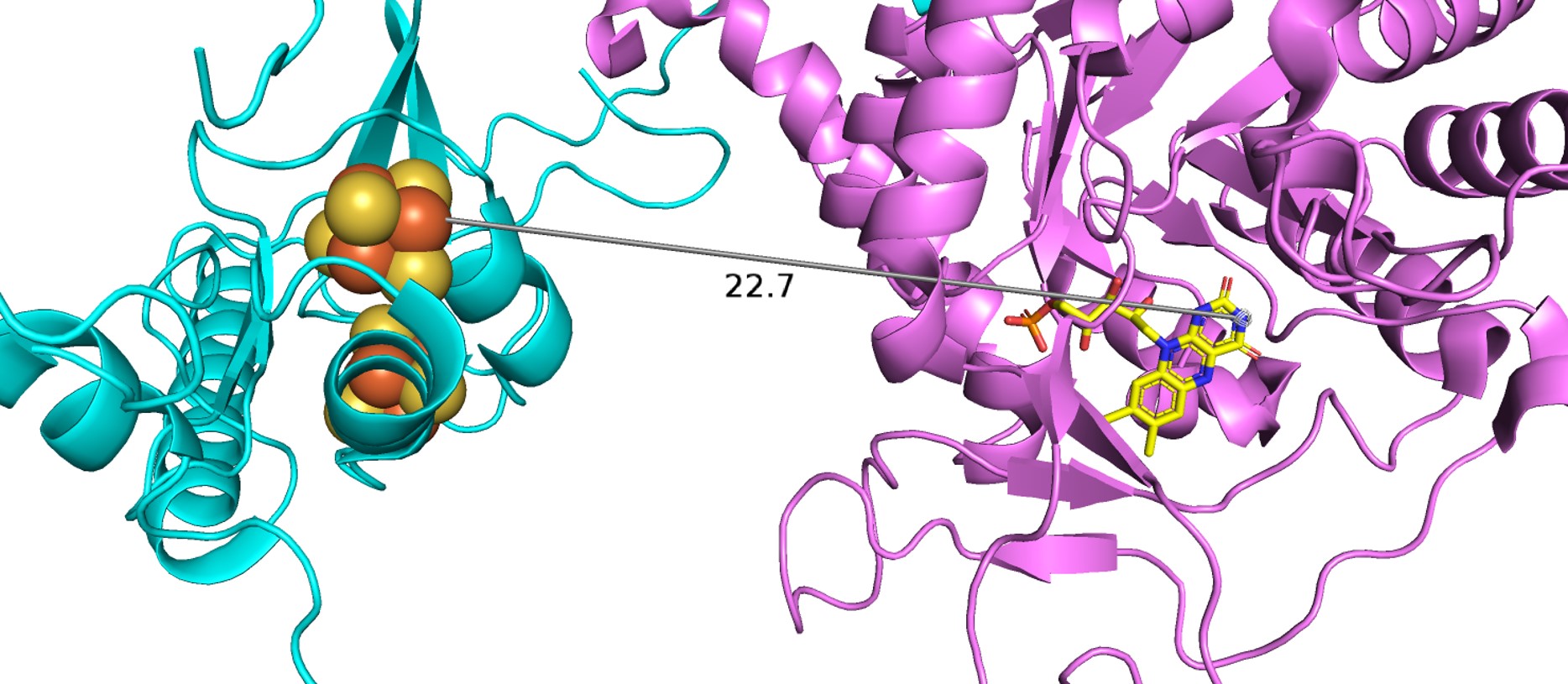
| + | Domain 5 consists of residues 1-26 and 848-1020. It also binds two [4Fe-4S] clusters. Between these clusters there are two α-helices and a four-stranded antiparallel β-sheet. The [4Fe-4S] clusters here in domian 5, which act as electron transfer centers, are not close to the next active redox site (the FMN binding site in domain 4). There is a gap of ~24 Å between N-terminal domain 5 Fe-S clusters and the FMN in the domian 4 binding site, which is too long for electrons to be able to cross. However, in dimer confirmation, the redox partners are close enough for electrons to transfer, thus, the enzyme must be in dimer confirmation to be active. | ||
| + | |||
| + | [[Image:D5.jpg]] | ||
| + | |||
| + | |||
| + | == Electron Transport Chain == | ||
| + | The enzyme has a two-site ping-pong mechanism with NADPH reducing FAD and reduced FMN responsible for reducing the pyrimidine. Transfer of electrons from NADPH to the pyrimidine is downhill with FMN rapidly reduced by FADH2 via the Fe-S conduit. Two single electrons are transferred via the Fe-S pathways. The reduction of the pyrimidine at the second active proceeds using general acid catalysis with protonation at N5 of FMN carried out by lysine-574, as FMN is reduced, and protonation at C5 of the pyrimidine by Cysteine-671, as it is reduced. | ||
| + | |||
| + | As earlier state, the dimer configuration is vitally important to the enzyme's function as electrons cannot travel through the Fe-S pathway without crossing over the dimer interface. In the isolated monomers, the gap between the N-terminal Fe-S clusters and FMN about 24 Å apart, too far for electrons to cross. As a result, all 12 redox co-factors of the DPD dimer are organized into two electron transfer chains passing the dimer interface twice. In this structural configuration, all redox partners are separated by ~7.5-10 Å, distances that are commonly observed in other multicenter electron transfer chains. Thus, DPD can only be active if it is in a dimer configuration and all of the Fe-S clusters participate in the electron transfer. | ||
| + | |||
| + | [[Image:etransport]] | ||
Revision as of 20:45, 13 December 2018
Structure of Eukaryotic Dihydropyrimidine Dehydrogenase
| |||||||||||
References
<Dobritzsch, D., Schneider, G., Schnackerz, K. D., & Lindqvist, Y. (2001). Crystal structure of dihydropyrimidine dehydrogenase, a major determinant of the pharmacokinetics of the anti‐cancer drug 5‐fluorouracil. The EMBO journal, 20(4), 650-660./> <Schnackerz, K. D., Dobritzsch, D., Lindqvist, Y., & Cook, P. F. (2004). Dihydropyrimidine dehydrogenase: a flavoprotein with four iron–sulfur clusters. Biochimica et Biophysica Acta (BBA)-Proteins and Proteomics, 1701(1-2), 61-74./>