Sandbox Reserved 1477
From Proteopedia
| Line 7: | Line 7: | ||
==Introduction== | ==Introduction== | ||
| - | The topic is | + | The topic is the study of the enzyme '''glucose oxidase''' (GOX) from '''aspergillus niger'''. It is not an enzyme from human body. Instead, it is found in the cells of fungus. These types of enzymes catalysts the oxidation of beta-D-glucose to δ-gluconolactone and H2O2 (which is not a reaction happening in human body) <ref>PMID:10216293</ref>. |
| - | Two PDB | + | GOX has a “considerable commercial importance” in biosensors according to the essay. This type of enzyme is wildly used in biosensor. Since it could react with glucose with a great selectivity and produce hydrogen peroxide, the amount of hydrogen peroxide is a function of the initial amount of the glucose ideally. For is reason, it is used for quantitively measuring the amount of concentration of glucose, or in a simpler way, measuring the concentration of blood glucose <ref>PMID: 24907743</ref>. Thus, it could be (actually had already been) widely used in quick blood glucose tests, which is essential to those who have diabetes. However, the usage of this enzyme is not exactly limited to the medical area. A good method of reflecting the concentration of glucose could also be used in food industry in measuring and controlling the sugar amount in food samples since most types of natural-sourced carbohydrate in food industry, such as sucrose and lactose, contain glucose <ref>PMID: 19374943</ref>. |
| + | |||
| + | Two PDB files mentioned in the essay, 1cf3 and 1gpe, which are two glucose oxidases from different organisms, ''aspergillus niger''' and '''Penicillium Amagasakiense'''. They have exactly the same types of ligands. The most important ligand is FAD cofactor, which assists the oxidation of beta-D-glucose. This essay was published back to 1999, which is fairly old in the scale of science. | ||
== Function == | == Function == | ||
Revision as of 02:32, 14 December 2018
| This Sandbox is Reserved from November 5 2018 through January 1, 2019 for use in the course "CHEM 4923: Senior Project taught by Christina R. Bourne at the University of Oklahoma, Norman, USA. This reservation includes Sandbox Reserved 1471 through Sandbox Reserved 1478. |
To get started:
More help: Help:Editing |
This page is reserved for Bo
Contents |
Default
This is a default text for your page '. Click above on edit this page' to modify. Be careful with the < and > signs. You may include any references to papers as in: the use of JSmol in Proteopedia [1] or to the article describing Jmol [2] to the rescue.
Introduction
The topic is the study of the enzyme glucose oxidase (GOX) from aspergillus niger. It is not an enzyme from human body. Instead, it is found in the cells of fungus. These types of enzymes catalysts the oxidation of beta-D-glucose to δ-gluconolactone and H2O2 (which is not a reaction happening in human body) [3].
GOX has a “considerable commercial importance” in biosensors according to the essay. This type of enzyme is wildly used in biosensor. Since it could react with glucose with a great selectivity and produce hydrogen peroxide, the amount of hydrogen peroxide is a function of the initial amount of the glucose ideally. For is reason, it is used for quantitively measuring the amount of concentration of glucose, or in a simpler way, measuring the concentration of blood glucose [4]. Thus, it could be (actually had already been) widely used in quick blood glucose tests, which is essential to those who have diabetes. However, the usage of this enzyme is not exactly limited to the medical area. A good method of reflecting the concentration of glucose could also be used in food industry in measuring and controlling the sugar amount in food samples since most types of natural-sourced carbohydrate in food industry, such as sucrose and lactose, contain glucose [5].
Two PDB files mentioned in the essay, 1cf3 and 1gpe, which are two glucose oxidases from different organisms, aspergillus niger' and Penicillium Amagasakiense. They have exactly the same types of ligands. The most important ligand is FAD cofactor, which assists the oxidation of beta-D-glucose. This essay was published back to 1999, which is fairly old in the scale of science.
Function
 The green part is the dimer of MAN, the blue part is the BMA (one of the reactants) and the yellow part is the a dimer of NAG.
Residues (the residues in red) were highlighted by the author because of their interaction with the ligands.
The green part is the dimer of MAN, the blue part is the BMA (one of the reactants) and the yellow part is the a dimer of NAG.
Residues (the residues in red) were highlighted by the author because of their interaction with the ligands.
Energetic
Structural highlights
|
This is a sample scene created with SAT to by Group, and another to make of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes.
The structure of this enzyme contains only one chain with 4 different types of ligands. This is a 583-residue-long protein with the molecular weight of about 65.8kDa. The structure is tested by X-ray diffraction for the certain PDB entry with the code 1cf3. X-ray diffraction gives the information of the whole protein sequence (if the resolution of the instrument is high enough), which makes it better than any other methods with non-100% sequence courage.
Another glucose oxidase also tested with X-ray diffraction method in the same essay from Penicillium Amagasakiense has two identical chains. As mentioned, they have exactly the same ligands, which actually make sense since they have the same function.
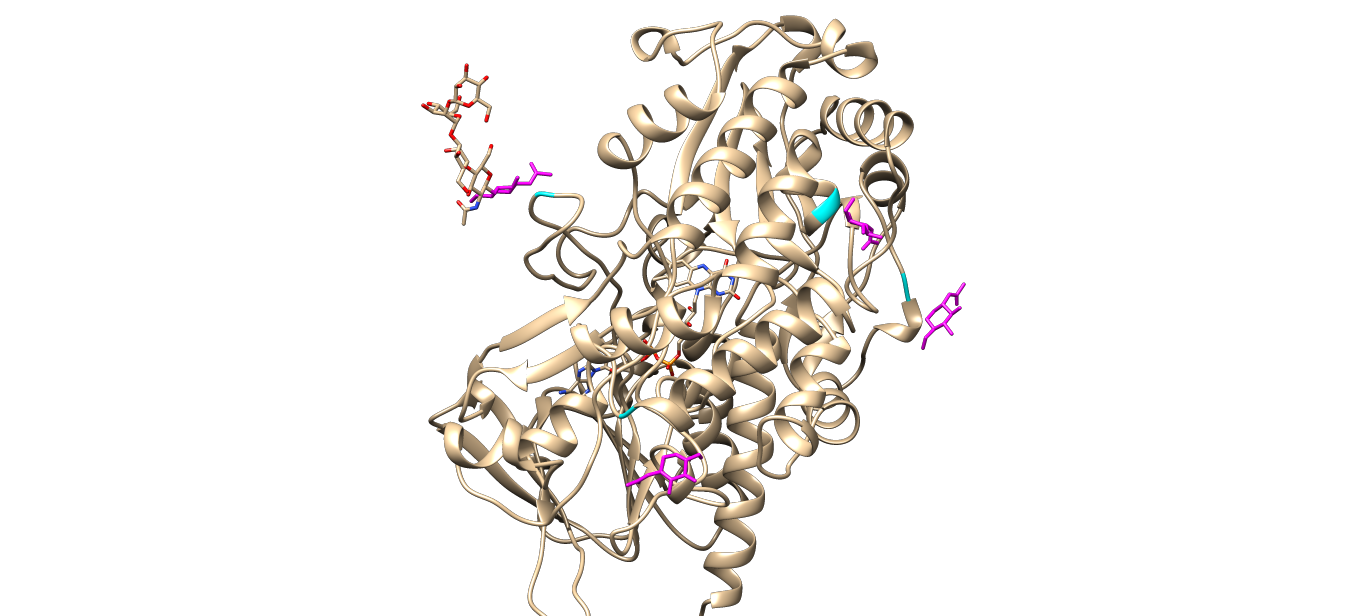 There are totally four NAG ligands (magenta part) around the enzyme which corresponding to the four ASN residues (cyan part) mentioned above, which might be the potential binding sites of the reactants.
There are totally four NAG ligands (magenta part) around the enzyme which corresponding to the four ASN residues (cyan part) mentioned above, which might be the potential binding sites of the reactants.
</StructureSection>
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ Wohlfahrt G, Witt S, Hendle J, Schomburg D, Kalisz HM, Hecht HJ. 1.8 and 1.9 A resolution structures of the Penicillium amagasakiense and Aspergillus niger glucose oxidases as a basis for modelling substrate complexes. Acta Crystallogr D Biol Crystallogr. 1999 May;55(Pt 5):969-77. PMID:10216293
- ↑ Tian K, Prestgard M, Tiwari A. A review of recent advances in nonenzymatic glucose sensors. Mater Sci Eng C Mater Biol Appl. 2014 Aug 1;41:100-18. doi:, 10.1016/j.msec.2014.04.013. Epub 2014 Apr 13. PMID:24907743 doi:http://dx.doi.org/10.1016/j.msec.2014.04.013
- ↑ Bankar SB, Bule MV, Singhal RS, Ananthanarayan L. Glucose oxidase--an overview. Biotechnol Adv. 2009 Jul-Aug;27(4):489-501. doi:, 10.1016/j.biotechadv.2009.04.003. Epub 2009 Apr 15. PMID:19374943 doi:http://dx.doi.org/10.1016/j.biotechadv.2009.04.003
