The human acetylcholinesterase (AChE) is an enzyme which hydrolyses the neurotransmitter acetylcholine (ACh) in the neuromuscular junctions and in other cholinergic synapses to terminate the neuronal signal.
It has an ellipsoidal shape with dimensions ~ 45Å x 60Å x 65Å. This protein is composed of 531 residues. It consists of 12-stranded, central mixed surrounded by 14 . It is a member of the α/β hydrolase fold family[1].
In the physiological conditions, AChE exists as tetramers associated with either collagen-like Q subunit (ColQ) or proline-rich membrane-anchoring protein (PRiMA). The AChE is linked with these anchoring molecules by a "tryptophan amphiphilic tetramerization" domain (WAT). There is also a monomeric form which is soluble in the blood.
The Active site gorge of AChE
The active site of AChE involves two sites: the peripheral site and the catalytic site.

The peripheral site is a transitional binding site of the substrate. It provides a region rich in aromatic amino acids that guide the ligands (ACh or other agonists) by setting an array of low-affinity binding sites. This hydrophobic region traps ACh and transfers it to the deep catalytic site.
The catalytic site of AChE consists of two subsites: the "esteratic" site and "the anionic" site.
In the "esteratic site" a catalytic triad consisting of forms a planar array that resembles the catalytic triad of serine proteases.
S203 is activated (it becomes nucleophilic) by E334 and H447. This activation allows the following reaction: the acylation between hydroxyl group of S203 and ACh oxygen (or other agonists). A covalent bond between the enzyme and the substrate creates an oxyanion. This oxyanion then reacts with two glycins setting up a hydrogen bond.
In the "anionic" site, the binds trimethylammonium group of ACh.
Further to these steps the substrate is well positioned to be hydrolysed into acetic acid and cholin.

Inhibitors of AChE
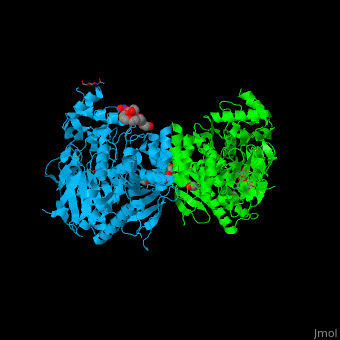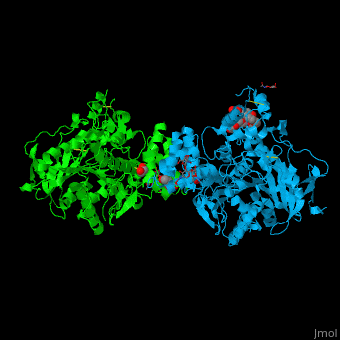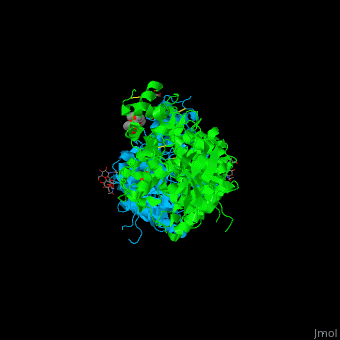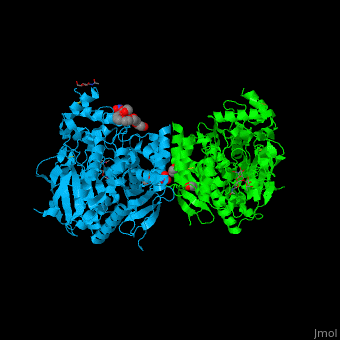
Fasciculin II
Fasciculin is a snake toxin. It is a little protein of 7kDa which inhibits AChE in binding the peripheric site, preventing the substrate from passing through the narrower portion of the gorge towards the catalytic site. This inhibition is almost irreversible.
The toxin is the one used in cristallisation of the Human acetylcholinesterase (in green on the picture).
Inhibitors used as treatments
We can find a lot of inhibitors such as in Alzheimer's disease drugs treatment. Actually, Alzheimer's disease is a neurodegenerative disease in which ACh is less present. An inhibition approach can be used to increase the remaining of ACh in the synaptic cleft by inhibiting the action of AChE. These treatments include rivastigmine, donepezil and tacrine. However, these drugs do not cure this disease, but only delay its development.
The molecule which has been the most studied is tacrine. A monomer of tacrine binds strongly to the peripheral site, preventing the subtrate from entry. When tacrine is in the dimer shape, it can bind the catalytic and peripheral sites of AChE.
Automated computational design of human enzymes for high bacterial expression and stability [2]
Upon heterologous overexpression, many proteins misfold or aggregate, thus resulting in low functional yields. Human acetylcholinesterase (hAChE), an enzyme mediating synaptic transmission, is a typical case of a human protein that necessitates mammalian systems to obtain functional expression. Using a novel computational strategy, we designed an AChE variant bearing 51 mutations that improved core packing, surface polarity, and backbone rigidity. This variant expressed at ~2,000-fold higher levels in E. coli compared to wild-type hAChE, and exhibited 20°C higher thermostability with no change in enzymatic properties or in the active-site configuration as determined by crystallography. To demonstrate broad utility, we similarly designed four other human and bacterial proteins. Testing at most three designs per protein, we obtained enhanced stability and/or higher yields of soluble protein in E. coli. Our algorithm requires only a 3D structure and several dozen sequences of naturally occurring homologues, and is available at http://pross.weizmann.ac.il.
. Wild type hAChE is shown in blue and 51 mutated positions, which are distributed throughout dAChE4, are indicated by orange spheres.
The choice of mutations at Gly416 in hAChE illustrates the role of these two filters (alignment scan and computational mutation scanning) in pruning false positives (see static image below). Position 416 is located on a partially exposed helical surface, where the small and flexible amino acid Gly is likely to destabilize hAChE. Indeed, in the alignment of 5 AChE homologues, Gly is infrequent and His is the most prevalent amino acid. Modeling shows, however, that in the specific context of hAChE, His adopts a strained side-chain conformation; in contrast, Gln, the third most prevalent amino acid, is predicted to be most stabilizing owing to its high helical propensity and favorable hydrogen-bonding with Tyr504. The combined filter therefore favors Gln over His for downstream design calculations.
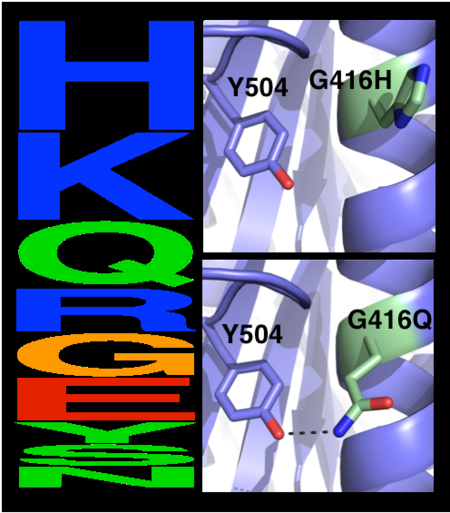
Eliminating potentially destabilizing mutations through homologous-sequence analysis and computational mutation scanning. Left: Sequence logo for hAChE position Gly416. The height of letters represents the respective amino acid’s frequency in an alignment of homologous AChE sequences. The evolutionarily ‘allowed’ sequence space (PSSM scores ≥0) at position 416 includes the 9 amino acids shown. Right: Structural models of mutations to the evolutionarily favored amino acid His, and to Gln, which is favored by Rosetta energy calculations. The His side chain is strained due to its proximity to the bulky Tyr504 aromatic ring, whereas the Gln side chain is relaxed and forms a favorable hydrogen bond with Tyr504 (dashed line)
Scenes highlight stabilizing effects of (in red), wild type hAChE is shown in cyan and designed hAChE is in green:
in the crystallographic structure of dAChE4 (PDB entry: 5hq3, yellow) compared to hAChE (PDB entry: 4ey4, green).
.
Comparison of the dAChE4 design model (yellow) with the solved crystal structure (PDB entry: 5hq3, green) and (wild-type hAChE (PDB entry: 4ey4, violet):
- .
- .
- in the model and structure is 3.1 Å (dashed line). This conformation change likely results from elimination of a side chain-backbone hydrogen bond between Thr112 and Ser110 due to the designed Thr112Ala mutation.
- . Val331Asn was predicted to form a hydrogen bond with Glu450 and another with Pro446 in the designed model; in the crystal structure, instead, Asn331 interacts with Glu334 and Glu450.
- .