Journal:Acta Cryst F:S2053230X18018083
From Proteopedia
(Difference between revisions)

| Line 7: | Line 7: | ||
[[Image:Figureacsf1.jpg|left|450px|thumb]] | [[Image:Figureacsf1.jpg|left|450px|thumb]] | ||
{{Clear}} | {{Clear}} | ||
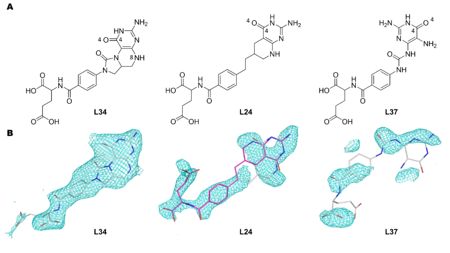
| - | Figure 1. The structures and names of DHCH inhibitors discussed in this work. A. L34 is (2S)-2-[[4-[(6aR)-3-amino-1,9-dioxo-5,6,6a,7-tetrahydro-4H-imidazo[1,5-f]pteridin-8-yl]benzoyl]amino]pentanedioic acid. L24 is (2R)-2-[[4-[2-[(6R)-2-amino-4-oxo-5,6,7,8-tetrahydro-1H-pyrido[2,3-d]pyrimidin-6-yl]ethyl]benzoyl]amino]pentanedioic acid. L37 is (2R)-2-[[4-[(2,5-diamino-4-oxo-1H-pyrimidin-6-yl)carbamoylamino]benzoyl]amino]pentanedioic acid. B. Omit Fo-Fc Polder maps (Liebschner et al., 2017) contoured at 3.0 r.m.s.d. after additional refinements. These omit maps were generated using the observed structure factors Fo of each model, and the calculated structure factors Fc generated after setting zero occupancies to the inhibitors, and removing the bulk solvent correction. The omit map for | + | '''Figure 1. The structures and names of DHCH inhibitors discussed in this work.''' '''A.''' L34 is (2S)-2-[[4-[(6aR)-3-amino-1,9-dioxo-5,6,6a,7-tetrahydro-4H-imidazo[1,5-f]pteridin-8-yl]benzoyl]amino]pentanedioic acid. L24 is (2R)-2-[[4-[2-[(6R)-2-amino-4-oxo-5,6,7,8-tetrahydro-1H-pyrido[2,3-d]pyrimidin-6-yl]ethyl]benzoyl]amino]pentanedioic acid. L37 is (2R)-2-[[4-[(2,5-diamino-4-oxo-1H-pyrimidin-6-yl)carbamoylamino]benzoyl]amino]pentanedioic acid. '''B.''' Omit Fo-Fc Polder maps (Liebschner ''et al''., 2017<ref name="Liebschner">PMID:28177311</ref>) contoured at 3.0 r.m.s.d. after additional refinements. These omit maps were generated using the observed structure factors Fo of each model, and the calculated structure factors Fc generated after setting zero occupancies to the inhibitors, and removing the bulk solvent correction. The omit map for [[1ecq]] is presented with the L24 pteridine ring in two orientations: white carbon sticks represents L24 pteridine ring in the same orientation as presented in [[1dia]] and pink carbon sticks represents the pteridine ring 180° rotated in relation to [[1dia]]. For L37, the coordinates from PDB entry [[1dia]] are shown. |
| - | One complex has a well-ordered ligand in a catalytic site and the model provides an improved description of enzyme-inhibitor interactions ([[1dib]]: <scene name='80/804503/Cv/2'>L34</scene>). One ligand may adopt two conformations in the binding site rather than the single one previously described ([[1dia]]: <scene name='80/804503/Cv/3'>L24</scene>; 1st conformation is colored in wheat and 2nd conformation is in | + | One complex has a well-ordered ligand in a catalytic site and the model provides an improved description of enzyme-inhibitor interactions ([[1dib]]: <scene name='80/804503/Cv/2'>L34</scene>). One ligand may adopt two conformations in the binding site rather than the single one previously described ([[1dia]]: <scene name='80/804503/Cv/3'>L24</scene>; 1st conformation is colored in wheat and 2nd conformation is in pink). There is no evidence to support incorporation of the third compound in the model ([[1dig]]:L37). Our interpretation of the data supports a correlation between the models and inhibition activity for two of the compounds. In the case of the third, inconsistencies are noted that would need to be addressed by further work. |
Revision as of 10:33, 23 December 2018
| |||||||||||
This page complements a publication in scientific journals and is one of the Proteopedia's Interactive 3D Complement pages. For aditional details please see I3DC.