We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1481
From Proteopedia
(Difference between revisions)
| Line 15: | Line 15: | ||
'''Primary structure''' | '''Primary structure''' | ||
| - | This complex is composed of two differents proteins forming two distinct but linked | + | This complex is composed of two differents proteins forming two distinct but linked subunits of the enzymatic complex. Each of these two proteins is coded by one different gene. The mettl3 gene codes for the N6-adenosine-methyltransferase 70 kDa subunit, which is a 225 Amino acid chain, whereas the Mettl14 gene codes for the N6-adenosine-methyltransferase subunit METTL14, which is composed of 349 amino acids. |
| - | Each of these two proteins are corresponding to the A [https://www.ncbi.nlm.nih.gov/protein/5K7M_A]or the B chain [https://www.ncbi.nlm.nih.gov/protein/5K7M_B] of the complex. | + | Each of these two proteins are corresponding to the A [https://www.ncbi.nlm.nih.gov/protein/5K7M_A]or the B chain [https://www.ncbi.nlm.nih.gov/protein/5K7M_B] of the complex and are neccessary to the functionning of the whole complex. |
'''Secondary structure''' | '''Secondary structure''' | ||
| - | Both polypeptides METTL3 and METTL14 are able to form some secondary | + | Both polypeptides METTL3 and METTL14 are able to form some secondary structures thank to interchain and intrachain hydrogen bond formation. |
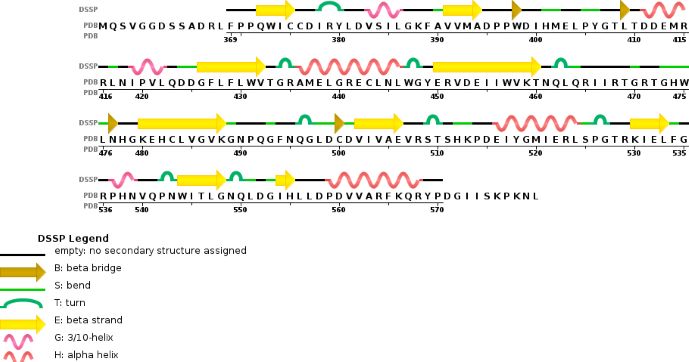
| - | Chain A METTL3'secondary structure, is mainly composed of 20% helical (7 helices; 46 residues)and 24% beta | + | Chain A METTL3'secondary structure, is mainly composed of 20% helical structures (7 helices; 46 residues) and 24% parallel and anti-parallel beta sheets (13 strands; 55 residues) |
[[Image:chain 1 METTL3.jpg]] | [[Image:chain 1 METTL3.jpg]] | ||
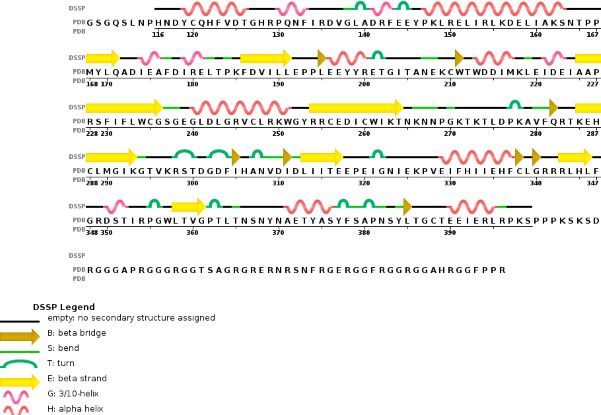
| - | Chain B METTL14 is made of 24% helical | + | Chain B METTL14 is made of 24% helical structures (14 helices; 87 residues) and 18% parallel and anti-parallel beta sheets (16 strands; 63 residues) |
[[Image:chain 2 METTL14.jpg]] | [[Image:chain 2 METTL14.jpg]] | ||
'''Tertiary structure''' | '''Tertiary structure''' | ||
| + | |||
'''Domains and specific site or sequences''' | '''Domains and specific site or sequences''' | ||
| Line 38: | Line 39: | ||
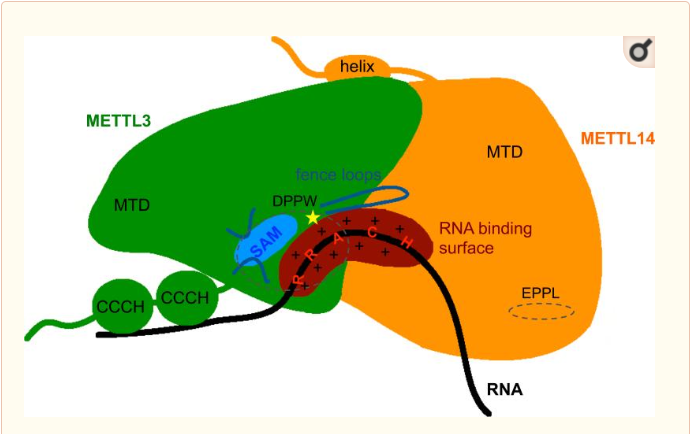
METTL3 and MTTL14 have both a methyltranferase domain but the complex METTL3/METTL14 has a better methyltransferase activity. Moreover, a mutatio in the catalytic center of METTL3 inhibits the hole methyltransferase activity of the complex, whereas a mutation in the catalytic center of METTL14 does not. Thus, METTL3 is the catalytic subunit of the complex and METTL14 enhances the methyltransfearse activity by stabilizing the complex and enables the recognition of consensus sequence on messenger RNA. | METTL3 and MTTL14 have both a methyltranferase domain but the complex METTL3/METTL14 has a better methyltransferase activity. Moreover, a mutatio in the catalytic center of METTL3 inhibits the hole methyltransferase activity of the complex, whereas a mutation in the catalytic center of METTL14 does not. Thus, METTL3 is the catalytic subunit of the complex and METTL14 enhances the methyltransfearse activity by stabilizing the complex and enables the recognition of consensus sequence on messenger RNA. | ||
| + | |||
| + | Zinc finger domain of the METTL3-METTL14 N6-methyladenosine methyltransferase, is a RNA binding domain of the complex. This perticular domain of the two proteins complex allow the protein to make interactions with the RNA molecule to modify | ||
[[Image:MET.PNG]] | [[Image:MET.PNG]] | ||
Revision as of 14:42, 29 December 2018
| This Sandbox is Reserved from 06/12/2018, through 30/06/2019 for use in the course "Structural Biology" taught by Bruno Kieffer at the University of Strasbourg, ESBS. This reservation includes Sandbox Reserved 1480 through Sandbox Reserved 1543. |
To get started:
More help: Help:Editing |
Crystal structure of the catalytic domains of Mettl3/Mettl14 complexInsert caption here
Drag the structure with the mouse to rotate
Insert caption here |
| Drag the structure with the mouse to rotate |
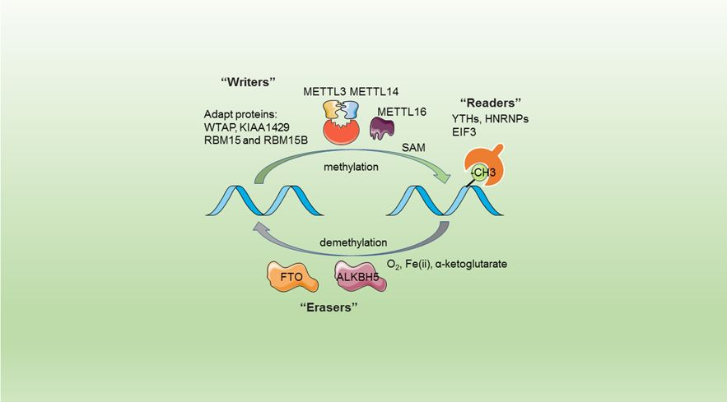
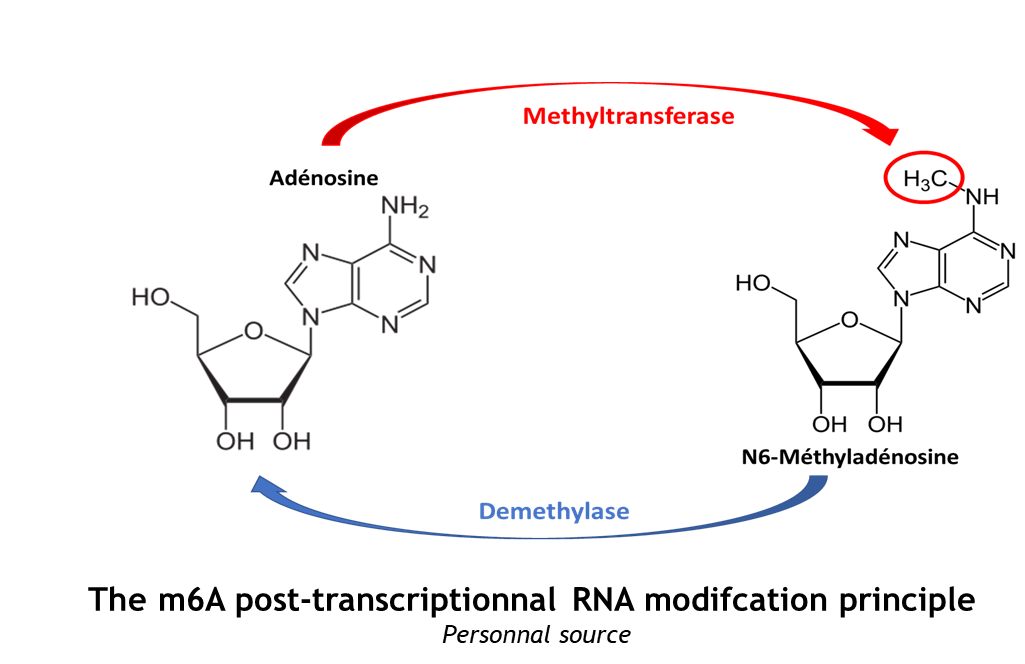
The complex METTL3/METTL14 is a heterodimer enzymatic complex involved into RNA post-transcription modifications by humans. This complex is abble to add a methyl group on adenosin of the RNA, by catalyzing a m6(A) modification.The N(6)-methyladenosine (m(6)A) is a quite common, reversible chemical modifications of RNAs molecules which plays a key role in several biological fonctions. This post transcriptional modification can be added by WRITERS, recognized by READERS and also removed byr ERASERS. The METTL3/METTL14 complex plays the role of writer.
| |||||||||||
References
- ↑ . PMID:216315890657
- ↑ . PMID:216315890657
- ↑ Wang P, Doxtader KA, Nam Y. Structural Basis for Cooperative Function of Mettl3 and Mettl14 Methyltransferases. Mol Cell. 2016 Jul 21;63(2):306-17. doi: 10.1016/j.molcel.2016.05.041. Epub 2016 , Jun 30. PMID:27373337 doi:http://dx.doi.org/10.1016/j.molcel.2016.05.041
- ↑ Wang X, Feng J, Xue Y, Guan Z, Zhang D, Liu Z, Gong Z, Wang Q, Huang J, Tang C, Zou T, Yin P. Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature. 2016 May 25;534(7608):575-8. doi: 10.1038/nature18298. PMID:27281194 doi:http://dx.doi.org/10.1038/nature18298
- ↑ Sledz P, Jinek M. Structural insights into the molecular mechanism of the m(6)A writer complex. Elife. 2016 Sep 14;5. pii: e18434. doi: 10.7554/eLife.18434. PMID:27627798 doi:http://dx.doi.org/10.7554/eLife.18434
- ↑ Wang X, Huang J, Zou T, Yin P. Human m(6)A writers: Two subunits, 2 roles. RNA Biol. 2017 Mar 4;14(3):300-304. doi: 10.1080/15476286.2017.1282025. Epub 2017, Jan 25. PMID:28121234 doi:http://dx.doi.org/10.1080/15476286.2017.1282025
- ↑ doi: https://dx.doi.org/10.2210/pdb5K7M/pdb