Sandbox Reserved 1481
From Proteopedia
| Line 1: | Line 1: | ||
{{Sandbox_Reserved_ESBS}}<!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | {{Sandbox_Reserved_ESBS}}<!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | ||
| - | ==Crystal structure of the catalytic domains of Mettl3/Mettl14 complex<Structure load='5K7M' size='350' frame='true' align='right' caption=' | + | ==Crystal structure of the catalytic domains of Mettl3/Mettl14 complex<Structure load='5K7M' size='350' frame='true' align='right' caption='Crystal structure of the catalytic domains of Mettl3/Mettl14 complex' scene='Insert optional scene name here' />== |
The complex METTL3/METTL14 is a heterodimer enzymatic complex involved into RNA post-transcriptional modifications by humans. | The complex METTL3/METTL14 is a heterodimer enzymatic complex involved into RNA post-transcriptional modifications by humans. | ||
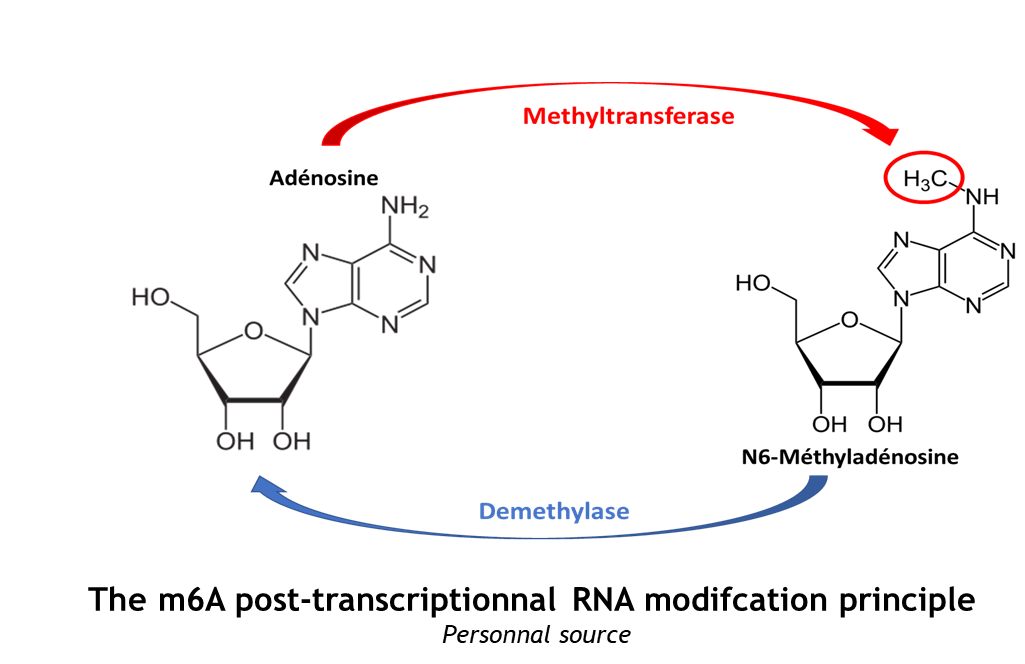
This complex is abble to add a methyl group on adenosin of the RNA, by catalyzing a m6(A) modification.The N(6)-methyladenosine (m(6)A) is a quite common, reversible chemical modification of RNAs molecules, which plays a key role in several different biological fonctions. This post-transcriptional modification can be added by WRITERS, recognized by READERS and also removed byr ERASERS. The METTL3/METTL14 complex plays the role of writer. | This complex is abble to add a methyl group on adenosin of the RNA, by catalyzing a m6(A) modification.The N(6)-methyladenosine (m(6)A) is a quite common, reversible chemical modification of RNAs molecules, which plays a key role in several different biological fonctions. This post-transcriptional modification can be added by WRITERS, recognized by READERS and also removed byr ERASERS. The METTL3/METTL14 complex plays the role of writer. | ||
| Line 44: | Line 44: | ||
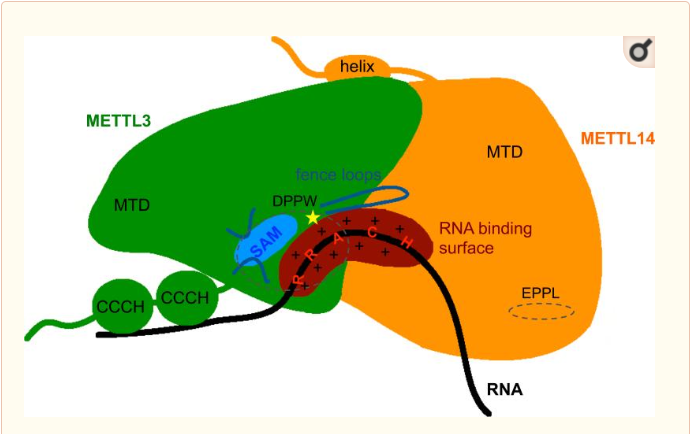
Thus, METTL3 is the catalytic subunit of the complex and METTL14 enhances the methyltransferasese activity by stabilizing the complex structure and binding to messenger RNA by enabling the recognition of its consensus sequence. | Thus, METTL3 is the catalytic subunit of the complex and METTL14 enhances the methyltransferasese activity by stabilizing the complex structure and binding to messenger RNA by enabling the recognition of its consensus sequence. | ||
| - | Both MTD have approximatly 25% sequence homology.Despite some common point the methyltransferase domain of METTL14 has extra terminal extensions, with an unusual N-terminal extension which is approximately 50 amino acid long. In the one hand the N-terminal extension create along helix which go through the domain and allow a close contact with the MTD of the catalytic subunit of the complex. This contact is made by several loops and shorter helical segments. In other hand, the C-terminal helix of the MTD of METTL14 is antiparallel to the N-terminal extension helix in order to stabilize its position. This allow the formation of a broad interdomain binding interface,and also others contact point along the terminal extensions, which are stabilizing the positions of both domains neccesary for the function.<Structure load='5YZ9' size='350' frame='true' align='right' caption=' | + | Both MTD have approximatly 25% sequence homology.Despite some common point the methyltransferase domain of METTL14 has extra terminal extensions, with an unusual N-terminal extension which is approximately 50 amino acid long. In the one hand the N-terminal extension create along helix which go through the domain and allow a close contact with the MTD of the catalytic subunit of the complex. This contact is made by several loops and shorter helical segments. In other hand, the C-terminal helix of the MTD of METTL14 is antiparallel to the N-terminal extension helix in order to stabilize its position. This allow the formation of a broad interdomain binding interface,and also others contact point along the terminal extensions, which are stabilizing the positions of both domains neccesary for the function.<Structure load='5YZ9' size='350' frame='true' align='right' caption='The zinc finger domain of METTL3 subunit' scene='Insert optional scene name here' /> |
| Line 60: | Line 60: | ||
METTL3 and METTL14 are linked to each other to form the functionnal complex. So as the METTL3 is responsible of the catalytic activity, the METTL14 allow the allosteric recognition of the RNA. The two subunit bindind creates a cavity in the center of the complex where the enzymatic reaction take place, it is the catalytic center. In these catalytic center some critical amino acid residues for the substrate recognition and specific binding are located. | METTL3 and METTL14 are linked to each other to form the functionnal complex. So as the METTL3 is responsible of the catalytic activity, the METTL14 allow the allosteric recognition of the RNA. The two subunit bindind creates a cavity in the center of the complex where the enzymatic reaction take place, it is the catalytic center. In these catalytic center some critical amino acid residues for the substrate recognition and specific binding are located. | ||
| - | == | + | == Crystal structure == |
| - | To study the | + | To study the crystal structure of the METTL3/METTL14 some crystalograghy experiments have been realized under the following experimental conditions to crystalyze the protein complex: |
'''Method''' : Vapor Diffusion Hanging Drop | '''Method''' : Vapor Diffusion Hanging Drop | ||
| Line 72: | Line 72: | ||
'''Buffer''': 0.1 M Tris pH 8.0, 20% PEG3350 | '''Buffer''': 0.1 M Tris pH 8.0, 20% PEG3350 | ||
| - | After | + | After crytalization, crystal had been analyzed through X-Ray diffraction, at 100°C, with a single wavelength coming out of a synchrotron. |
Thanks to the experiment the following data have been collected to describe the cristal structure of the complex : | Thanks to the experiment the following data have been collected to describe the cristal structure of the complex : | ||
| Line 87: | Line 87: | ||
[[Image:Cristallo.jpeg]] | [[Image:Cristallo.jpeg]] | ||
| - | The | + | The crystal structure analysis at 1,65 angstrom allow to collect precise and detailled informations about the whole structure like the side chains and also the hydrogen bonds. ''(Cristal analysis data at other resolution are available on [http://www.rcsb.org/pdb/explore/materialsAndMethods.do?structureId=5K7M].)'' |
Revision as of 16:40, 29 December 2018
| This Sandbox is Reserved from 06/12/2018, through 30/06/2019 for use in the course "Structural Biology" taught by Bruno Kieffer at the University of Strasbourg, ESBS. This reservation includes Sandbox Reserved 1480 through Sandbox Reserved 1543. |
To get started:
More help: Help:Editing |
Crystal structure of the catalytic domains of Mettl3/Mettl14 complexCrystal structure of the catalytic domains of Mettl3/Mettl14 complex
Drag the structure with the mouse to rotate
Crystal structure of the catalytic domains of Mettl3/Mettl14 complex |
| Drag the structure with the mouse to rotate |
The complex METTL3/METTL14 is a heterodimer enzymatic complex involved into RNA post-transcriptional modifications by humans. This complex is abble to add a methyl group on adenosin of the RNA, by catalyzing a m6(A) modification.The N(6)-methyladenosine (m(6)A) is a quite common, reversible chemical modification of RNAs molecules, which plays a key role in several different biological fonctions. This post-transcriptional modification can be added by WRITERS, recognized by READERS and also removed byr ERASERS. The METTL3/METTL14 complex plays the role of writer.
This enzymatic complex belongs to the second class of enzyme, which are the transferases. The complex is formed by 574 amino acid residues, divided into two different proteins nammed as Methyltransferase Like number 3 and 14. 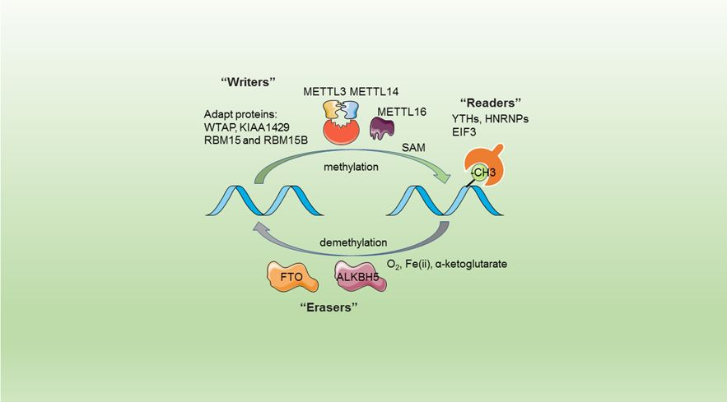
| |||||||||||
References
- ↑ . PMID:216315890657
- ↑ . PMID:216315890657
- ↑ Wang P, Doxtader KA, Nam Y. Structural Basis for Cooperative Function of Mettl3 and Mettl14 Methyltransferases. Mol Cell. 2016 Jul 21;63(2):306-17. doi: 10.1016/j.molcel.2016.05.041. Epub 2016 , Jun 30. PMID:27373337 doi:http://dx.doi.org/10.1016/j.molcel.2016.05.041
- ↑ Wang X, Feng J, Xue Y, Guan Z, Zhang D, Liu Z, Gong Z, Wang Q, Huang J, Tang C, Zou T, Yin P. Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature. 2016 May 25;534(7608):575-8. doi: 10.1038/nature18298. PMID:27281194 doi:http://dx.doi.org/10.1038/nature18298
- ↑ Sledz P, Jinek M. Structural insights into the molecular mechanism of the m(6)A writer complex. Elife. 2016 Sep 14;5. pii: e18434. doi: 10.7554/eLife.18434. PMID:27627798 doi:http://dx.doi.org/10.7554/eLife.18434
- ↑ Wang X, Huang J, Zou T, Yin P. Human m(6)A writers: Two subunits, 2 roles. RNA Biol. 2017 Mar 4;14(3):300-304. doi: 10.1080/15476286.2017.1282025. Epub 2017, Jan 25. PMID:28121234 doi:http://dx.doi.org/10.1080/15476286.2017.1282025
- ↑ doi: https://dx.doi.org/10.2210/pdb5K7M/pdb