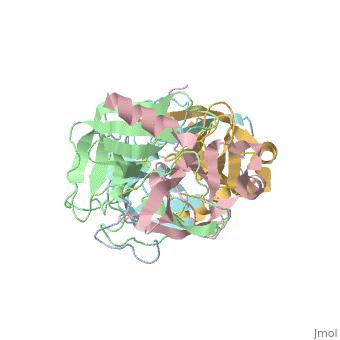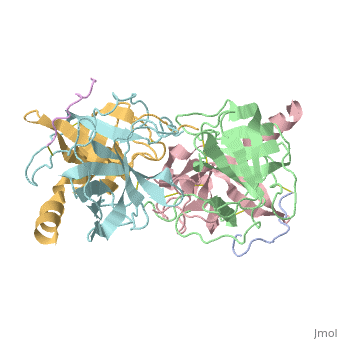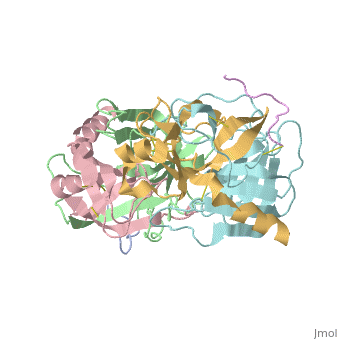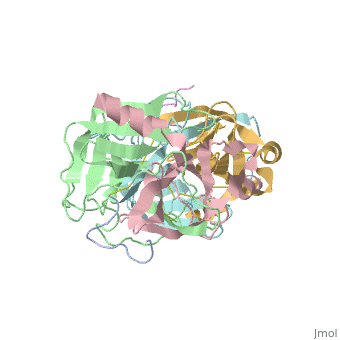4cha
From Proteopedia
| Line 5: | Line 5: | ||
|SITE= | |SITE= | ||
|LIGAND= | |LIGAND= | ||
| - | |ACTIVITY= [http://en.wikipedia.org/wiki/Chymotrypsin Chymotrypsin], with EC number [http://www.brenda-enzymes.info/php/result_flat.php4?ecno=3.4.21.1 3.4.21.1] | + | |ACTIVITY= <span class='plainlinks'>[http://en.wikipedia.org/wiki/Chymotrypsin Chymotrypsin], with EC number [http://www.brenda-enzymes.info/php/result_flat.php4?ecno=3.4.21.1 3.4.21.1] </span> |
|GENE= | |GENE= | ||
| + | |DOMAIN= | ||
| + | |RELATEDENTRY= | ||
| + | |RESOURCES=<span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=4cha FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=4cha OCA], [http://www.ebi.ac.uk/pdbsum/4cha PDBsum], [http://www.rcsb.org/pdb/explore.do?structureId=4cha RCSB]</span> | ||
}} | }} | ||
| Line 27: | Line 30: | ||
[[Category: hydrolase (serine proteinase)]] | [[Category: hydrolase (serine proteinase)]] | ||
| - | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on | + | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Mon Mar 31 05:38:15 2008'' |
Revision as of 02:38, 31 March 2008
| |||||||
| , resolution 1.68Å | |||||||
|---|---|---|---|---|---|---|---|
| Activity: | Chymotrypsin, with EC number 3.4.21.1 | ||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||
STRUCTURE OF ALPHA-*CHYMOTRYPSIN REFINED AT 1.68 ANGSTROMS RESOLUTION
Overview
Diffraction data for alpha-chymotrypsin crystals at -10 degrees C were measured at 1.68 A resolution and refined by restrained structure-factor least-squares refinement. The two independent chymotrypsin molecules in the crystallographic asymmetric unit were refined independently. The overall structure of alpha-chymotrypsin is little changed from published co-ordinates. The root-mean-square shift of C alpha co-ordinates is 0.42 A, co-ordinates for the two molecules showing a root-mean-square difference of 0.19 A. Certain regions with high disorder (residues 9 to 14, 73 to 79) remain difficult to interpret and several side-chains are disordered. Some water molecule positions have been changed. The absence of the tosyl group has made a significant difference to the refined structure at the active site. This now agrees closely with other enzymes of the trypsin family that have been refined at high resolution. There is a strong hydrogen bond between N epsilon 2 (His57) and O gamma (Ser195) in the free enzyme, in line with the published description of the charge relay system.
About this Structure
4CHA is a Protein complex structure of sequences from Bos taurus. Full crystallographic information is available from OCA.
Reference
Structure of alpha-chymotrypsin refined at 1.68 A resolution., Tsukada H, Blow DM, J Mol Biol. 1985 Aug 20;184(4):703-11. PMID:4046030
Page seeded by OCA on Mon Mar 31 05:38:15 2008