We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1489
From Proteopedia
(Difference between revisions)
| Line 7: | Line 7: | ||
== '''Function''' == | == '''Function''' == | ||
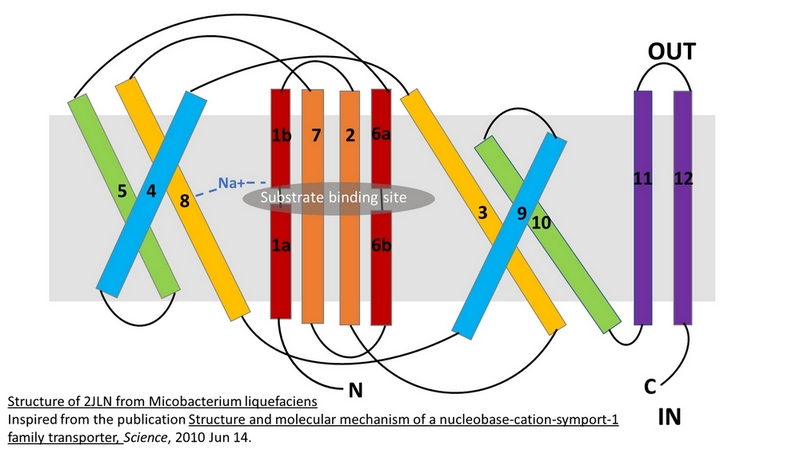
| - | Mhp1 is a | + | Mhp1 is a trans-membrane protein bellowing to the nucleobase-cation-symport-1 (NCS1) transporter family from Microbacterium liquefaciens. |
| + | |||
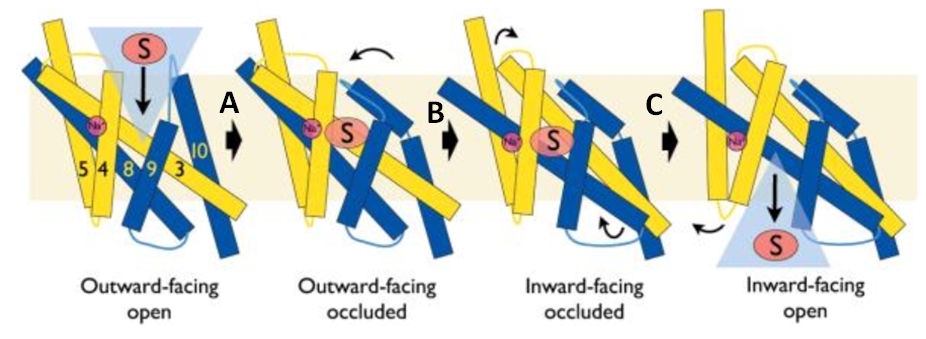
| + | A transporter effects the cellular uptake and release of a wide range of substances across biological membranes in all organisms. This is done by coupling the uphill movement of the substrate against its concentration gradient with the energetically favorable downhill gradient of a second substrate, often a proton or a cation. The kinetics and thermodynamics of the transporters can be explained by their alternating conformations.[doi: 10.1126/science.1186303] | ||
| + | |||
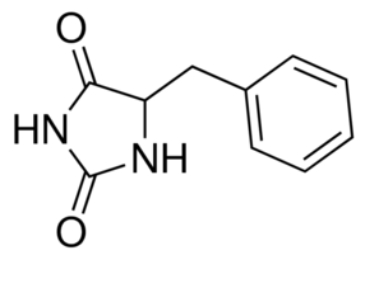
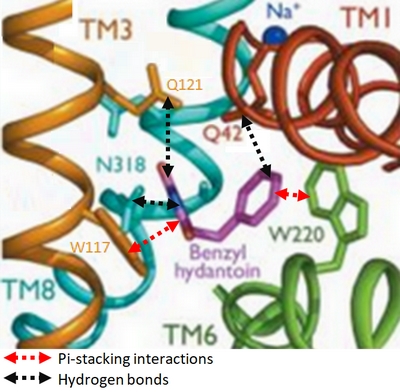
| + | In the case of Mhp1, it allows the sodium dependent income of indolyl methyl- and benzyl-hydantoins (''Figure 1'') in the cell. Those are part of a salvage metabolic pathway leading to their conversion in amino acids. | ||
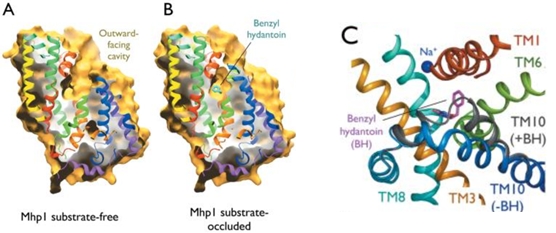
2JLN is one of the conformations of Mhp1. It is the outward-facing conformation without substrate. | 2JLN is one of the conformations of Mhp1. It is the outward-facing conformation without substrate. | ||
| Line 13: | Line 17: | ||
[[Image:Structure of benzyl-hydantoin.jpg]] | [[Image:Structure of benzyl-hydantoin.jpg]] | ||
| - | ''Figure 1 : Structure of benzyl-hydantoin'' | + | ''Figure 1 : Structure of benzyl-hydantoin, the substrate of Mhp1'' |
Revision as of 22:59, 9 January 2019
| This Sandbox is Reserved from 06/12/2018, through 30/06/2019 for use in the course "Structural Biology" taught by Bruno Kieffer at the University of Strasbourg, ESBS. This reservation includes Sandbox Reserved 1480 through Sandbox Reserved 1543. |
To get started:
More help: Help:Editing |
2JLN
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644