We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1491
From Proteopedia
(Difference between revisions)
| Line 60: | Line 60: | ||
The development of cancer begins with the modification of the sequence and expression of the genes involved in the [https://en.wikipedia.org/wiki/Cell_cycle cell cycle]. <ref>https://en.wikipedia.org/wiki/Cancer</ref> | The development of cancer begins with the modification of the sequence and expression of the genes involved in the [https://en.wikipedia.org/wiki/Cell_cycle cell cycle]. <ref>https://en.wikipedia.org/wiki/Cancer</ref> | ||
| + | |||
The transformation from healthy cells to cancer cells is carried out in two stages: carcinogenesis and tumorigenesis. | The transformation from healthy cells to cancer cells is carried out in two stages: carcinogenesis and tumorigenesis. | ||
During [https://en.wikipedia.org/wiki/Carcinogenesis carcinogenesis], cells accumulate genetic abnormalities, particularly in [https://en.wikipedia.org/wiki/Oncogene oncogenic] sequences. Oncogenes are positive regulators of cell proliferation. After a mutation, they become hyperactive and cause an excessive cellular growth. [https://en.wikipedia.org/wiki/Caretaker_gene Gatekeeper genes] (genes that allow the passage from one stage of the cell cycle to the next) can also be mutated, leading to uncontrolled cell proliferation. | During [https://en.wikipedia.org/wiki/Carcinogenesis carcinogenesis], cells accumulate genetic abnormalities, particularly in [https://en.wikipedia.org/wiki/Oncogene oncogenic] sequences. Oncogenes are positive regulators of cell proliferation. After a mutation, they become hyperactive and cause an excessive cellular growth. [https://en.wikipedia.org/wiki/Caretaker_gene Gatekeeper genes] (genes that allow the passage from one stage of the cell cycle to the next) can also be mutated, leading to uncontrolled cell proliferation. | ||
During tumorigenesis, cancer becomes invasive: cancer cells invade other healthy organs. | During tumorigenesis, cancer becomes invasive: cancer cells invade other healthy organs. | ||
| + | |||
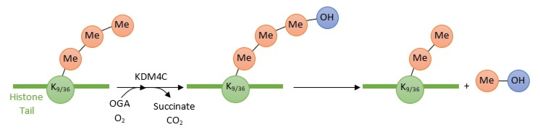
KDM4C is involved in carcinogenesis as an oncogene. Indeed, by catalyzing the demethylation of H3K9-me3 (lysine 9 from trimethylated histones 3) to H3K9-me2 (lysine 9 from dimethylated histones 3), this protein increases the expression of its target genes. Several KDM4C target genes are involved in cell growth. For example, they influence mitogenic signalling - which promotes mitosis and cell division -, cell cycle regulation and translation. | KDM4C is involved in carcinogenesis as an oncogene. Indeed, by catalyzing the demethylation of H3K9-me3 (lysine 9 from trimethylated histones 3) to H3K9-me2 (lysine 9 from dimethylated histones 3), this protein increases the expression of its target genes. Several KDM4C target genes are involved in cell growth. For example, they influence mitogenic signalling - which promotes mitosis and cell division -, cell cycle regulation and translation. | ||
| - | In cancer cells, KDM4C expression is enhanced. Thus, the growth of tumor cells is greatly increased. | + | In cancer cells, KDM4C expression is enhanced. Thus, the growth of tumor cells is greatly increased. <ref>Gregory, Brittany L., and Vivian G. Cheung. ‘Natural Variation in the Histone Demethylase, KDM4C, Influences Expression Levels of Specific Genes Including Those That Affect Cell Growth’. Genome Research 24, no. 1 (January 2014): 52–63. https://doi.org/10.1101/gr.156141.113</ref> |
| - | In addition, KDM4C is involved in the correct segregation of chromosomes. Its high presence in tumor cells therefore ensures their viability. | + | |
| + | |||
| + | In addition, KDM4C is involved in the correct segregation of chromosomes. Its high presence in tumor cells therefore ensures their viability.<ref>Garcia, Jeison, and Fernando Lizcano. ‘KDM4C Activity Modulates Cell Proliferation and Chromosome Segregation in Triple-Negative Breast Cancer’. Breast Cancer : Basic and Clinical Research 10 (2 November 2016): 169–75. https://doi.org/10.4137/BCBCR.S40182.</ref> | ||
| + | |||
Finally, KDM4C also plays a role in the tumorigenesis of certain cancers, such as breast cancer, since it allows the proliferation of cancer cells, their migration and their invasive capacity in the triple-negative breast cancer. | Finally, KDM4C also plays a role in the tumorigenesis of certain cancers, such as breast cancer, since it allows the proliferation of cancer cells, their migration and their invasive capacity in the triple-negative breast cancer. | ||
| + | |||
For all the implications of KDM4C in different cancers, it is one of the main targets of anti-cancer treatments. | For all the implications of KDM4C in different cancers, it is one of the main targets of anti-cancer treatments. | ||
Revision as of 15:10, 10 January 2019
| This Sandbox is Reserved from 06/12/2018, through 30/06/2019 for use in the course "Structural Biology" taught by Bruno Kieffer at the University of Strasbourg, ESBS. This reservation includes Sandbox Reserved 1480 through Sandbox Reserved 1543. |
To get started:
More help: Help:Editing |
2xml
Preview
2xml is a 2 chain structure. This domain belongs to the Human KDM4C protein.
KDM4C is a histone demethylase involved in the specific demethylation of trimethylated residues (Lys 9 and Lys 36 of histone 3). These marks are specific tags for epigenetic activation. KDM4C plays a main role in the modification of cell cycle genes expression and thus involved in the growth of tumoral cells.
| |||||||||||
References
- ↑ http://consurf.tau.ac.il/fgij/fg.htm?mol=/temp/2XMLA_ConSurf_DB_pipe.pdb
- ↑ Douglas Hanahan et Robert A. Weinberg, « The hallmarks of cancer », Cell, vol. 100, 7 janvier 2000, p. 57-70 (PMID 10647931)
- ↑ https://en.wikipedia.org/wiki/Cancer
- ↑ Gregory, Brittany L., and Vivian G. Cheung. ‘Natural Variation in the Histone Demethylase, KDM4C, Influences Expression Levels of Specific Genes Including Those That Affect Cell Growth’. Genome Research 24, no. 1 (January 2014): 52–63. https://doi.org/10.1101/gr.156141.113
- ↑ Garcia, Jeison, and Fernando Lizcano. ‘KDM4C Activity Modulates Cell Proliferation and Chromosome Segregation in Triple-Negative Breast Cancer’. Breast Cancer : Basic and Clinical Research 10 (2 November 2016): 169–75. https://doi.org/10.4137/BCBCR.S40182.