Sandbox Reserved 1491
From Proteopedia
(Difference between revisions)
| Line 13: | Line 13: | ||
== Function == | == Function == | ||
| - | '''Genes expression''' is directly related to the '''condensation state''' of the [https://en.wikipedia.org/wiki/Chromatin chromatin]. Indeed, chromatin can be in the form of [https://en.wikipedia.org/wiki/Heterochromatin heterochromatin] (condensed form of DNA) or [https://en.wikipedia.org/wiki/Euchromatin euchromatin] (relaxed form of DNA) which correspond respectively to the transcriptionally silent and active forms of DNA. Chromatin is composed of DNA wrapped around [https://en.wikipedia.org/wiki/Histone histone] octamers forming nucleosomes. The '''histone tails residues''' can be [https://en.wikipedia.org/wiki/Acetylation acetylated], [https://en.wikipedia.org/wiki/Methylation methylated] or [https://en.wikipedia.org/wiki/Demethylation demethylated] by enzymes in order to modify chromatin state and therefore gene expression. Different types of proteins involved in this process exist, such as histone | + | '''Genes expression''' is directly related to the '''condensation state''' of the [https://en.wikipedia.org/wiki/Chromatin chromatin]. Indeed, chromatin can be in the form of [https://en.wikipedia.org/wiki/Heterochromatin heterochromatin] (condensed form of DNA) or [https://en.wikipedia.org/wiki/Euchromatin euchromatin] (relaxed form of DNA) which correspond respectively to the transcriptionally silent and active forms of DNA. Chromatin is composed of DNA wrapped around [https://en.wikipedia.org/wiki/Histone histone] octamers forming nucleosomes. The '''histone tails residues''' can be [https://en.wikipedia.org/wiki/Acetylation acetylated], [https://en.wikipedia.org/wiki/Methylation methylated] or [https://en.wikipedia.org/wiki/Demethylation demethylated] by enzymes in order to modify chromatin state and therefore gene expression. Different types of proteins involved in this process exist, such as histone acetylase (HAT), histone methylase (HMT) or histone demethylase (HDM). |
Two families of [https://en.wikipedia.org/wiki/Demethylase histone-lysine demethylase] (KDM) have been identified as follows : the '''flavin (FAD)-dependent lysine-specific demethylases''' and the '''Fe(II)-dependent Jumonji C (JmjC) family'''. JmjC is subfamily of histone demethylases which regroups several proteins containing a specific catalytic domain called '''Jmjc''' found in ''' 2xml structure'''. KDM4 demethylases belong to the JmjC family and contains six members : KDM4A-F | Two families of [https://en.wikipedia.org/wiki/Demethylase histone-lysine demethylase] (KDM) have been identified as follows : the '''flavin (FAD)-dependent lysine-specific demethylases''' and the '''Fe(II)-dependent Jumonji C (JmjC) family'''. JmjC is subfamily of histone demethylases which regroups several proteins containing a specific catalytic domain called '''Jmjc''' found in ''' 2xml structure'''. KDM4 demethylases belong to the JmjC family and contains six members : KDM4A-F | ||
| Line 19: | Line 19: | ||
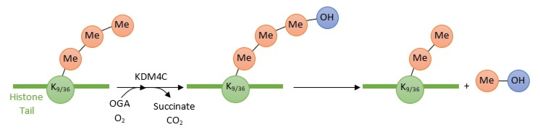
[[Image:Reactionjpg.jpg | thumb | upright=3 | Enzymatic reaction of demethylation of H3K9(me3) and H3K36(me3) by KDM4C ]] | [[Image:Reactionjpg.jpg | thumb | upright=3 | Enzymatic reaction of demethylation of H3K9(me3) and H3K36(me3) by KDM4C ]] | ||
'''KDM4C/JMJD2''' is a protein which converts specific trimethylated histone residues to the dimethylated form. Indeed, it catalyzes the demethylation of both '''Lysine 9 and Lysine 36 of histone 3''' (respectively H3K9me3 and H3K36me3 by hydroxylation of the lysine methyl group. This reaction leads to a dissociation of the methyl group from the lysine histone tail. KDM4C employs (OG), Fe2+ and oxygen as cosubstrates to promote its enzymatic reaction, thus the '''dissociation of methyl groups'''. | '''KDM4C/JMJD2''' is a protein which converts specific trimethylated histone residues to the dimethylated form. Indeed, it catalyzes the demethylation of both '''Lysine 9 and Lysine 36 of histone 3''' (respectively H3K9me3 and H3K36me3 by hydroxylation of the lysine methyl group. This reaction leads to a dissociation of the methyl group from the lysine histone tail. KDM4C employs (OG), Fe2+ and oxygen as cosubstrates to promote its enzymatic reaction, thus the '''dissociation of methyl groups'''. | ||
| + | |||
| + | |||
== Structural highlights == | == Structural highlights == | ||
| Line 49: | Line 51: | ||
== Disease == | == Disease == | ||
| - | [https://en.wikipedia.org/wiki/Cancer Cancer] is a disease characterized by an abnormally high level of cell proliferation, known as a [https://en.wikipedia.org/wiki/Neoplasm tumor]. This uncontrolled growth is the result of a modification of genetic information and its expression. | + | [https://en.wikipedia.org/wiki/Cancer '''Cancer'''] is a disease characterized by an abnormally high level of cell proliferation, known as a [https://en.wikipedia.org/wiki/Neoplasm tumor]. This uncontrolled growth is the result of a '''modification of genetic information and its expression'''. |
Cancer cells have several characteristics, related to the activity of KDM4C: | Cancer cells have several characteristics, related to the activity of KDM4C: | ||
| Line 61: | Line 63: | ||
The development of cancer begins with the modification of the sequence and expression of the genes involved in the [https://en.wikipedia.org/wiki/Cell_cycle cell cycle]. <ref>https://en.wikipedia.org/wiki/Cancer</ref> | The development of cancer begins with the modification of the sequence and expression of the genes involved in the [https://en.wikipedia.org/wiki/Cell_cycle cell cycle]. <ref>https://en.wikipedia.org/wiki/Cancer</ref> | ||
| - | The transformation from healthy cells to cancer cells is carried out in two stages: carcinogenesis and tumorigenesis. | + | The transformation from healthy cells to cancer cells is carried out in two stages: '''carcinogenesis and tumorigenesis'''. |
| - | During [https://en.wikipedia.org/wiki/Carcinogenesis carcinogenesis], cells accumulate genetic abnormalities, particularly in [https://en.wikipedia.org/wiki/Oncogene oncogenic] sequences. Oncogenes are positive regulators of cell proliferation. After a mutation, they become hyperactive and cause an excessive cellular growth. [https://en.wikipedia.org/wiki/Caretaker_gene Gatekeeper genes] (genes that allow the passage from one stage of the cell cycle to the next) can also be mutated, leading to uncontrolled cell proliferation. | + | During [https://en.wikipedia.org/wiki/Carcinogenesis carcinogenesis], cells accumulate '''genetic abnormalities''', particularly in [https://en.wikipedia.org/wiki/Oncogene oncogenic] sequences. '''Oncogenes''' are positive regulators of cell proliferation. After a mutation, they become hyperactive and cause an excessive cellular growth. [https://en.wikipedia.org/wiki/Caretaker_gene '''Gatekeeper genes'''] (genes that allow the passage from one stage of the cell cycle to the next) can also be mutated, leading to uncontrolled cell proliferation. |
During tumorigenesis, cancer becomes invasive: cancer cells invade other healthy organs. | During tumorigenesis, cancer becomes invasive: cancer cells invade other healthy organs. | ||
| Line 72: | Line 74: | ||
In addition, KDM4C is involved in the correct segregation of chromosomes. Its high presence in tumor cells therefore ensures their viability.<ref>Garcia, Jeison, and Fernando Lizcano. ‘KDM4C Activity Modulates Cell Proliferation and Chromosome Segregation in Triple-Negative Breast Cancer’. Breast Cancer : Basic and Clinical Research 10 (2 November 2016): 169–75. https://doi.org/10.4137/BCBCR.S40182.</ref> | In addition, KDM4C is involved in the correct segregation of chromosomes. Its high presence in tumor cells therefore ensures their viability.<ref>Garcia, Jeison, and Fernando Lizcano. ‘KDM4C Activity Modulates Cell Proliferation and Chromosome Segregation in Triple-Negative Breast Cancer’. Breast Cancer : Basic and Clinical Research 10 (2 November 2016): 169–75. https://doi.org/10.4137/BCBCR.S40182.</ref> | ||
| - | Finally, KDM4C also plays a role in the tumorigenesis of certain cancers, such as breast cancer, since it allows the proliferation of cancer cells, their migration and their invasive capacity in the triple-negative breast cancer. | + | Finally, KDM4C also plays a role in the tumorigenesis of certain cancers, such as '''breast cancer''', since it allows the proliferation of cancer cells, their migration and their invasive capacity in the triple-negative breast cancer. |
| - | For all the implications of KDM4C in different cancers, it is one of the main targets of anti-cancer treatments. | + | For all the implications of KDM4C in different cancers, it is one of the main '''targets of anti-cancer treatments'''. |
Revision as of 15:15, 10 January 2019
| This Sandbox is Reserved from 06/12/2018, through 30/06/2019 for use in the course "Structural Biology" taught by Bruno Kieffer at the University of Strasbourg, ESBS. This reservation includes Sandbox Reserved 1480 through Sandbox Reserved 1543. |
To get started:
More help: Help:Editing |
2xml
Preview
2xml is a 2 chain structure. This domain belongs to the Human KDM4C protein.
KDM4C is a histone demethylase involved in the specific demethylation of trimethylated residues (Lys 9 and Lys 36 of histone 3). These marks are specific tags for epigenetic activation. KDM4C plays a main role in the modification of cell cycle genes expression and thus involved in the growth of tumoral cells.
| |||||||||||
References
- ↑ http://consurf.tau.ac.il/fgij/fg.htm?mol=/temp/2XMLA_ConSurf_DB_pipe.pdb
- ↑ Douglas Hanahan et Robert A. Weinberg, « The hallmarks of cancer », Cell, vol. 100, 7 janvier 2000, p. 57-70 (PMID 10647931)
- ↑ https://en.wikipedia.org/wiki/Cancer
- ↑ Gregory, Brittany L., and Vivian G. Cheung. ‘Natural Variation in the Histone Demethylase, KDM4C, Influences Expression Levels of Specific Genes Including Those That Affect Cell Growth’. Genome Research 24, no. 1 (January 2014): 52–63. https://doi.org/10.1101/gr.156141.113
- ↑ Garcia, Jeison, and Fernando Lizcano. ‘KDM4C Activity Modulates Cell Proliferation and Chromosome Segregation in Triple-Negative Breast Cancer’. Breast Cancer : Basic and Clinical Research 10 (2 November 2016): 169–75. https://doi.org/10.4137/BCBCR.S40182.