We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1490
From Proteopedia
(Difference between revisions)
| Line 2: | Line 2: | ||
==Kinase Domain of Tyrosine-protein kinase receptor TIE-2 (PDB:6MWE)== | ==Kinase Domain of Tyrosine-protein kinase receptor TIE-2 (PDB:6MWE)== | ||
<StructureSection load='6MWE' size='340' side='right' caption='Caption for this structure' scene='' name='pp'> | <StructureSection load='6MWE' size='340' side='right' caption='Caption for this structure' scene='' name='pp'> | ||
| - | The protein we are focusing one is a protein kinase receptor to a family of ligands called angiopoietins. This receptor is a Tyrosine Kinase TIE2. We are going to analyze the kinase domain of this protein | ||
| + | The protein we are focusing one is a protein kinase receptor to a family of ligands called angiopoietins. This receptor is a Tyrosine Kinase TIE2. | ||
It acts as cell-surface receptor for the ligands ANGPT1, ANGPT2 and ANGPT4 and regulates among others angiogenesis, endothelial cell survival and maintenance of vascular quiescence. It is important in the regulation of both normal physiologic and pathologic angiogenesis. The later is a fundamental step in the transition of tumors from a benign state to a malignant one. | It acts as cell-surface receptor for the ligands ANGPT1, ANGPT2 and ANGPT4 and regulates among others angiogenesis, endothelial cell survival and maintenance of vascular quiescence. It is important in the regulation of both normal physiologic and pathologic angiogenesis. The later is a fundamental step in the transition of tumors from a benign state to a malignant one. | ||
| + | Angiogenesis is the process in which new blood vessels are formed from pre-existing blood vessels. The growth of these new blood vessels requires migration and proliferation of endothelial cells (ECs). It is an event controlled by angiogenic growth factors such as vascular endothelial growth factor (VEGF). | ||
| + | While ANGPT1 is a TIE2 agonist and has a higher binding affinity to it than ANGPT2, ANGPT2 can act as a context-dependent agonist. Thus, the ANGPT/TIE2 kinase signaling pathway is an attractive anti-vascular target. | ||
| - | Angiogenesis is the process in which new blood vessels are formed from pre-existing blood vessels. The growth of these new blood vessels requires migration and proliferation of endothelial cells (ECs). It is an event controlled by angiogenic growth factors such as vascular endothelial growth factor (VEGF). | ||
| - | |||
| - | While ANGPT1 is a TIE2 agonist and has a higher binding affinity to it than ANGPT2, ANGPT2 can act as a context-dependent agonist. Thus, the ANGPT/TIE2 kinase signaling pathway is an attractive anti-vascular target. | ||
Be careful with the < and > signs. | Be careful with the < and > signs. | ||
| Line 14: | Line 13: | ||
== Function == | == Function == | ||
| - | |||
| - | |||
| - | |||
== Structural highlights == | == Structural highlights == | ||
| Line 29: | Line 25: | ||
Arginine at position 849 is found in six residues upstream of the invariant lysine K855 in the kinase domain (sequence preserved among the human, bovine, murine and rat TIE2 sequences). This seems to prove that a basic amino acid is essential for this position. In addition, arginine located a few amino acids before invariant lysine is involved in stabilizing the kinase domain (hydrogen binding of arginine with a proline downstream). It is therefore possible that R849 may also be involved in the stabilization of the kinase domain. Thus, the substitution of R849 by a W could modify the conformation of the kinase domain, leading to a decrease in inhibitory mechanisms and involving autophosphorylation. | Arginine at position 849 is found in six residues upstream of the invariant lysine K855 in the kinase domain (sequence preserved among the human, bovine, murine and rat TIE2 sequences). This seems to prove that a basic amino acid is essential for this position. In addition, arginine located a few amino acids before invariant lysine is involved in stabilizing the kinase domain (hydrogen binding of arginine with a proline downstream). It is therefore possible that R849 may also be involved in the stabilization of the kinase domain. Thus, the substitution of R849 by a W could modify the conformation of the kinase domain, leading to a decrease in inhibitory mechanisms and involving autophosphorylation. | ||
| - | + | With this mutation, Venous Malformations (VMs) contain a Disproportionately high ratio of Endothelial Cells (ECs) to Smooth Muscle Cells (SMCs) | |
| - | ( | + | |
[[Image:Venous Malformations Diagram.jpg]] | [[Image:Venous Malformations Diagram.jpg]] | ||
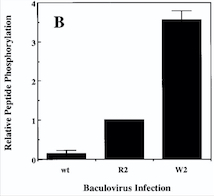
| + | ''Fig 1 : Comparison of the Kinase Activities of Normal and Mutant TIE2 Receptors'' | ||
| + | ''(B) Cells infected with wild-type baculovirus (wt) or virus expressing normal TIE2 (R2) or mutant TIE2 (W2). Cells expressing the mutation at position 849 (Arginine → Tryptophan) have an autophosphorylation activity 6 to 10 times higher than wild cells.'' | ||
| + | [[Image:Venous Malformations Immunohistochemistry.jpg]] | ||
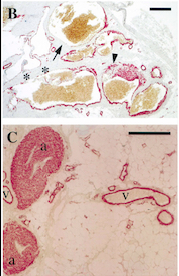
| + | ''Fig 2 : Immunohistochemistry of VMs with Antibodies against Smooth Muscle Cells 𝛂-Actin'' | ||
| - | + | ''B = Abnormal channels'' | |
| - | '' | + | ''C = Normal veins (v) and arteries (a)'' |
| - | + | ''Scale bars, 200 𝛍m'' | |
| - | + | ''Antibodies directed against SMCs 𝛂-Actin from cells with VMs show that the vessels have a specific and abnormal staining (B) compared to normal vessels (C)'' | |
| - | Scale bars, 200 𝛍m. | ||
| - | Antibodies directed against SMCs 𝛂-Actin from cells with VMs show that the vessels have a specific and abnormal staining (B) compared to normal vessels (C) | ||
| - | |||
| - | [[Image:Venous Malformations Immunohistochemistry.jpg]] | ||
== Relevance == | == Relevance == | ||
Revision as of 15:42, 10 January 2019
| This Sandbox is Reserved from 06/12/2018, through 30/06/2019 for use in the course "Structural Biology" taught by Bruno Kieffer at the University of Strasbourg, ESBS. This reservation includes Sandbox Reserved 1480 through Sandbox Reserved 1543. |
To get started:
More help: Help:Editing |
Kinase Domain of Tyrosine-protein kinase receptor TIE-2 (PDB:6MWE)
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644