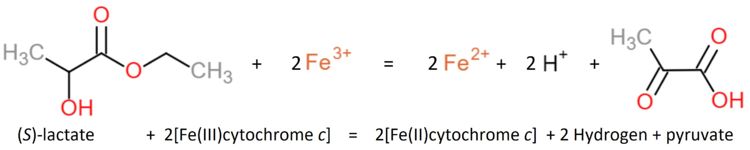We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1501
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
{{Sandbox_Reserved_ESBS}}<!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | {{Sandbox_Reserved_ESBS}}<!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | ||
<StructureSection load='1stp' size='340' side='right' caption='Caption for this structure' scene=''> | <StructureSection load='1stp' size='340' side='right' caption='Caption for this structure' scene=''> | ||
| - | This is a default text for your page ''''''. Click above on '''edit this page''' to modify. | ||
| - | You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | ||
| - | |||
Encoded in the ''Saccharomyces cerevisiae'' (strain ATCC 204508 / S288c) gene CYB2 the protein Flavocytochrome b(2) | Encoded in the ''Saccharomyces cerevisiae'' (strain ATCC 204508 / S288c) gene CYB2 the protein Flavocytochrome b(2) | ||
It is an Oxidoreductase categorized according to IUB as EC: 1.1.2.3. | It is an Oxidoreductase categorized according to IUB as EC: 1.1.2.3. | ||
| Line 15: | Line 12: | ||
[[Image:Reaction1.JPG|center|x150px]] | [[Image:Reaction1.JPG|center|x150px]] | ||
| - | Similar oxidants that can be used to perform the reaction in vitro are ferricyanide, phenozine methosulfate and quinone [experiment first performed 1963 by Nygaard, later in 1966 described by Symons and Burgoyne]. | + | Similar oxidants that can be used to perform the reaction in vitro are ferricyanide, phenozine methosulfate and quinone [experiment first performed 1963 by Nygaard, later in 1966 described by Symons and Burgoyne]<ref>DOI 10.1002/ijch.201300024</ref>. |
| - | The Yeasts L-Lactate Dehydrogenase can be inhibited by heavy metals, oxygen, glycerate, oxalate, malate, phenylpyruvate and fatty acids [Nygaard 1963]. | + | The Yeasts L-Lactate Dehydrogenase can be inhibited by heavy metals, oxygen, glycerate, oxalate, malate, phenylpyruvate and fatty acids [Nygaard 1963]<ref>PMID:21638687</ref>. |
The Enzyme shows a specificity for L-lactate but none for the D-isomer or -hydroxybutyrate. | The Enzyme shows a specificity for L-lactate but none for the D-isomer or -hydroxybutyrate. | ||
Revision as of 16:56, 10 January 2019
| This Sandbox is Reserved from 06/12/2018, through 30/06/2019 for use in the course "Structural Biology" taught by Bruno Kieffer at the University of Strasbourg, ESBS. This reservation includes Sandbox Reserved 1480 through Sandbox Reserved 1543. |
To get started:
More help: Help:Editing |
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644