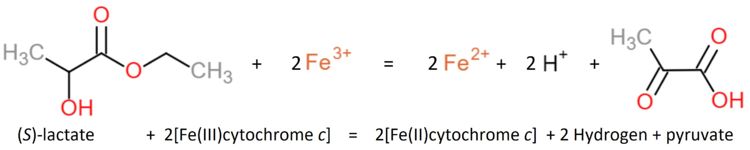We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1501
From Proteopedia
(Difference between revisions)
| Line 18: | Line 18: | ||
<!-- Global Stoichiometry Homotetramer A4 --> | <!-- Global Stoichiometry Homotetramer A4 --> | ||
| - | Flavocytochrome b(2) is a tetrameric enzyme (Jacq and Lederer, 1972 and 1974<ref> | + | Flavocytochrome b(2) is a tetrameric enzyme (Jacq and Lederer, 1972 and 1974<ref>PMID: 4336855</ref>,<ref>PMID: 4152980</ref>). Each of the four identical subunits is composed by one single polypeptide chain. |
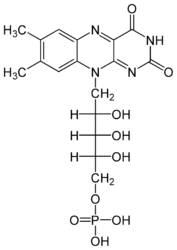
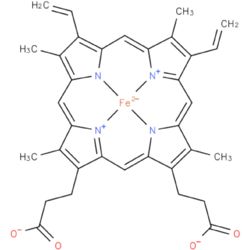
Each subunit contains a binding site for the selectively non-covalently binding of the cofactor FMN(3-) (Flavinmononucleotide), as well as one in with the iron complexed in the tetrapyrrole ring interacts with heme b(2-) cofactor (Risler and Groudinsky, 1973<ref>PMID: 5545004 </ref>). | Each subunit contains a binding site for the selectively non-covalently binding of the cofactor FMN(3-) (Flavinmononucleotide), as well as one in with the iron complexed in the tetrapyrrole ring interacts with heme b(2-) cofactor (Risler and Groudinsky, 1973<ref>PMID: 5545004 </ref>). | ||
| Line 24: | Line 24: | ||
<!-- https://www.ebi.ac.uk/chebi/searchId.do?chebiId=CHEBI:60344--> | <!-- https://www.ebi.ac.uk/chebi/searchId.do?chebiId=CHEBI:60344--> | ||
| - | The amino acid sequence in the heme binding region was first determined by Guidard ''et al'', 1974<ref> | + | The amino acid sequence in the heme binding region was first determined by Guidard ''et al'', 1974<ref>PMID: 4575975</ref>. |
For every subunit of the wild type protein form, the crystallized preparation analysis determined a molecular weight of the chain of 36 kD (Appleby and Morton, 1959<ref>PMID: 13638255</ref>) and the chain of 21 kD (Jacq and Lederer, 1974<ref>PMID: 4152980</ref>). | For every subunit of the wild type protein form, the crystallized preparation analysis determined a molecular weight of the chain of 36 kD (Appleby and Morton, 1959<ref>PMID: 13638255</ref>) and the chain of 21 kD (Jacq and Lederer, 1974<ref>PMID: 4152980</ref>). | ||
The sulfite adduct recombinant enzyme produced when expressed in ''E. coli'' was also crystallized (Tegoni and Cambillau, 1994<ref>PMID: 8003966</ref>) so key active site residues could be identified and comparisons with the mutant protein. | The sulfite adduct recombinant enzyme produced when expressed in ''E. coli'' was also crystallized (Tegoni and Cambillau, 1994<ref>PMID: 8003966</ref>) so key active site residues could be identified and comparisons with the mutant protein. | ||
Revision as of 19:03, 10 January 2019
| This Sandbox is Reserved from 06/12/2018, through 30/06/2019 for use in the course "Structural Biology" taught by Bruno Kieffer at the University of Strasbourg, ESBS. This reservation includes Sandbox Reserved 1480 through Sandbox Reserved 1543. |
To get started:
More help: Help:Editing |
| |||||||||||
References
- ↑ NYGAARD AP. Various forms of D- and L-lactate dehydrogenases in yeast. Ann N Y Acad Sci. 1961 Nov 2;94:774-9. PMID:14480786
- ↑ doi: https://dx.doi.org/10.1016/0076-6879(66)09064-5
- ↑ NYGAARD AP. Various forms of D- and L-lactate dehydrogenases in yeast. Ann N Y Acad Sci. 1961 Nov 2;94:774-9. PMID:14480786
- ↑ Jacq C, Lederer F. [Two molecular species of cytochrome b 2 from Saccharomyces cerevisiae]. Eur J Biochem. 1972 Jan 31;25(1):41-8. PMID:4336855
- ↑ Lederer F. On the first steps of lactate oxidation by bakers' yeast L-(plus)-lactate dehydrogenase (cytochrome b2). Eur J Biochem. 1974 Jul 15;46(2):393-9. PMID:4152980
- ↑ Groudinsky O. Study of heme-protein linkage in cytochrome b2. Destruction of a crucial histidine residue by photooxidation of "apo" cytochrome b2 core in the presence of rose bengal. Eur J Biochem. 1971 Feb;18(4):480-4. PMID:5545004
- ↑ Guiard B, Groudinsky O, Lederer F. Yeast L-lactate dehydrogenase (cytochrome b 2 ). Chemical characterization of the heme-binding core. Eur J Biochem. 1973 Apr;34(2):241-7. PMID:4575975
- ↑ APPLEBY CA, MORTON RK. Lactic dehydrogenase and cytochrome b2 of baker's yeast; purification and crystallization. Biochem J. 1959 Mar;71(3):492-9. PMID:13638255
- ↑ Lederer F. On the first steps of lactate oxidation by bakers' yeast L-(plus)-lactate dehydrogenase (cytochrome b2). Eur J Biochem. 1974 Jul 15;46(2):393-9. PMID:4152980
- ↑ Tegoni M, Cambillau C. The 2.6-A refined structure of the Escherichia coli recombinant Saccharomyces cerevisiae flavocytochrome b2-sulfite complex. Protein Sci. 1994 Feb;3(2):303-13. PMID:8003966
- ↑ Tegoni M, Cambillau C. The 2.6-A refined structure of the Escherichia coli recombinant Saccharomyces cerevisiae flavocytochrome b2-sulfite complex. Protein Sci. 1994 Feb;3(2):303-13. PMID:8003966