We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1489
From Proteopedia
(Difference between revisions)
| Line 47: | Line 47: | ||
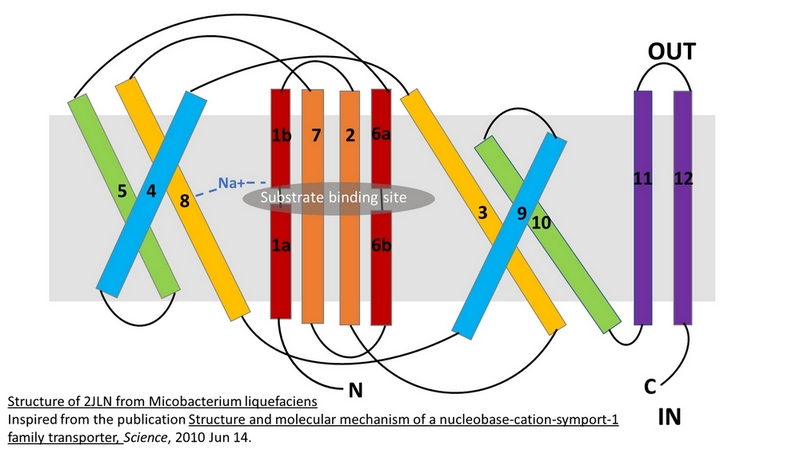
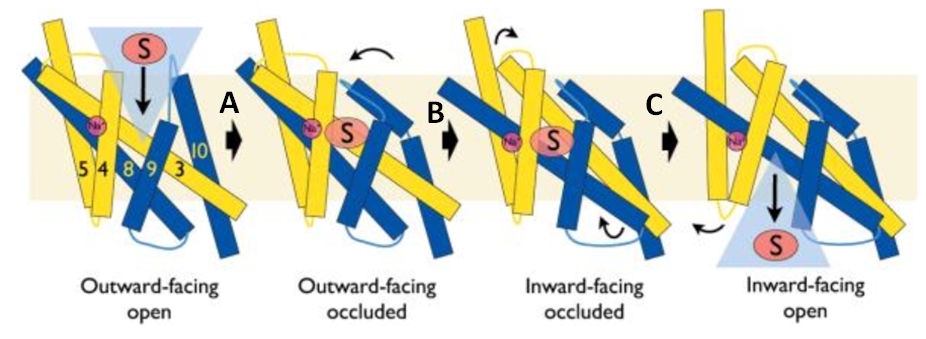
The central bundle is composed of TMs 1 and 2, twined to the TMs 6 and 7 respectively. In addition, the protein presents a V-shape structure formed by TMs 3 to 5, twined to TMs 8 to 10 (''Figure 2''). | The central bundle is composed of TMs 1 and 2, twined to the TMs 6 and 7 respectively. In addition, the protein presents a V-shape structure formed by TMs 3 to 5, twined to TMs 8 to 10 (''Figure 2''). | ||
| - | TM5 and TM10 are 'flexible helices' because they bend during the state transitions.<ref | + | TM5 and TM10 are 'flexible helices' because they bend during the state transitions.<ref name="art2" /> |
The substrate- and cation-binding sites are located in the space between the central four-helix-bundle and the outer helix layer. | The substrate- and cation-binding sites are located in the space between the central four-helix-bundle and the outer helix layer. | ||
| Line 70: | Line 70: | ||
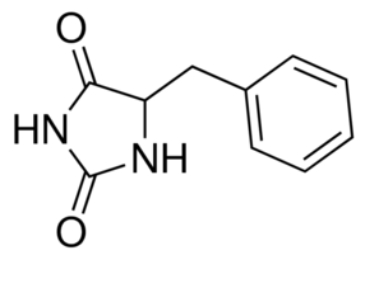
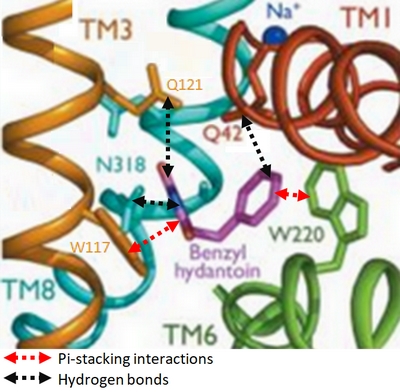
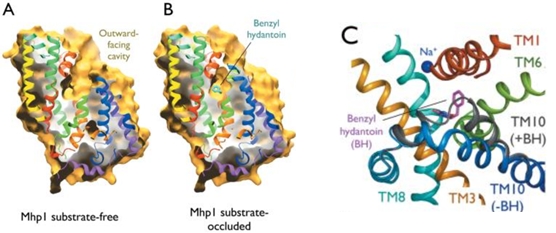
Experiments have shown that sodium increases the affinity of benzyl-hydantoin for Mhp1 and reciprocally benzyl-hydantoin increases the affinity of sodium for Mhp1. | Experiments have shown that sodium increases the affinity of benzyl-hydantoin for Mhp1 and reciprocally benzyl-hydantoin increases the affinity of sodium for Mhp1. | ||
| - | Indeed, the presence of benzyl-hydantoin in Mhp1 binding site blocks the pathway of the sodium ion to the extracellular side.<ref | + | Indeed, the presence of benzyl-hydantoin in Mhp1 binding site blocks the pathway of the sodium ion to the extracellular side.<ref name="art2" /> |
Therefore, the binding of the substrate and the cation are closely coupled. | Therefore, the binding of the substrate and the cation are closely coupled. | ||
Revision as of 20:24, 10 January 2019
| This Sandbox is Reserved from 06/12/2018, through 30/06/2019 for use in the course "Structural Biology" taught by Bruno Kieffer at the University of Strasbourg, ESBS. This reservation includes Sandbox Reserved 1480 through Sandbox Reserved 1543. |
To get started:
More help: Help:Editing |
2JLN
| |||||||||||
References
- ↑ Weyand S, Shimamura T, Yajima S, Suzuki S, Mirza O, Krusong K, Carpenter EP, Rutherford NG, Hadden JM, O'Reilly J, Ma P, Saidijam M, Patching SG, Hope RJ, Norbertczak HT, Roach PC, Iwata S, Henderson PJ, Cameron AD. Structure and Molecular Mechanism of a Nucleobase-Cation-Symport-1 Family Transporter. Science. 2008 Oct 16. PMID:18927357
- ↑ 2.0 2.1 2.2 2.3 Shimamura T, Weyand S, Beckstein O, Rutherford NG, Hadden JM, Sharples D, Sansom MS, Iwata S, Henderson PJ, Cameron AD. Molecular basis of alternating access membrane transport by the sodium-hydantoin transporter Mhp1. Science. 2010 Apr 23;328(5977):470-3. PMID:20413494 doi:328/5977/470
- ↑ Simmons KJ, Jackson SM, Brueckner F, Patching SG, Beckstein O, Ivanova E, Geng T, Weyand S, Drew D, Lanigan J, Sharples DJ, Sansom MS, Iwata S, Fishwick CW, Johnson AP, Cameron AD, Henderson PJ. Molecular mechanism of ligand recognition by membrane transport protein, Mhp1. EMBO J. 2014 Jun 21. pii: e201387557. PMID:24952894 doi:http://dx.doi.org/10.15252/embj.201387557