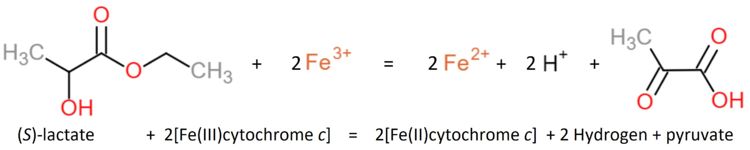This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Sandbox Reserved 1501
From Proteopedia
(Difference between revisions)
| Line 25: | Line 25: | ||
The amino acid sequence in the heme binding region was first determined by Guidard ''et al'', 1974<ref>PMID: 4210211</ref>. | The amino acid sequence in the heme binding region was first determined by Guidard ''et al'', 1974<ref>PMID: 4210211</ref>. | ||
| - | For every subunit of the wild type protein form, the crystallized preparation analysis determined a molecular weight of the chain of 36 kD (Appleby and Morton, 1959<ref>PMID: 13638255</ref>) and the chain of 21 kD (Jacq and Lederer, 1974<ref>PMID: 4152980</ref>). | + | For every subunit of the wild type protein form, the crystallized preparation analysis determined a molecular weight of the α-chain of 36 kD (Appleby and Morton, 1959<ref>PMID: 13638255</ref>) and the β-chain of 21 kD (Jacq and Lederer, 1974<ref>PMID: 4152980</ref>). |
| - | The sulfite adduct recombinant enzyme produced when expressed in ''E. coli'' was also crystallized (Tegoni and Cambillau, 1994<ref name="bob">PMID: 8003966</ref>) so key active site residues could be identified and comparisons with the mutant protein. | + | The sulfite adduct recombinant enzyme produced when expressed in ''E. coli'' was also crystallized (Tegoni and Cambillau, 1994<ref name="bob">PMID: 8003966</ref>) so key active site residues could be identified and comparisons with the mutant protein can be made. |
| - | The Arg289 of the ''E. coli'' wild type sulfite adduct can adopt two different conformations, one of which its side chain is stacked against Arg376, that directly interacts with the substrate, while in the second one the Arg289 side chain points towards the active site. | + | The Arg289 of the ''E. coli'' wild type sulfite adduct can adopt two different conformations, in one of which its side chain is stacked against Arg376, that directly interacts with the substrate, while in the second one the Arg289 side chain points towards the active site. |
The mutation changes that Arg289 into a Lysine becoming R289K-b(2). The mutant ARG289LYS can still be found in both conformations but it is now changing the kinetics of the reactions (Tegoni and Cambillau, 1994<ref name="bob"/>). | The mutation changes that Arg289 into a Lysine becoming R289K-b(2). The mutant ARG289LYS can still be found in both conformations but it is now changing the kinetics of the reactions (Tegoni and Cambillau, 1994<ref name="bob"/>). | ||
It is rising the K<sub>i</sub> of several components in comparison to the wild type, while k<sub>cat</sub> and K<sub>M</sub> are also changed by a factor of 10. | It is rising the K<sub>i</sub> of several components in comparison to the wild type, while k<sub>cat</sub> and K<sub>M</sub> are also changed by a factor of 10. | ||
It changes also the induction by L-lactate. Patterns of inhibition by pyruvate and oxalate are altered and the enzyme stops being inhibited by substrate excess. | It changes also the induction by L-lactate. Patterns of inhibition by pyruvate and oxalate are altered and the enzyme stops being inhibited by substrate excess. | ||
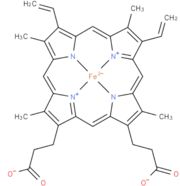
| - | The mutation has altered the flavin reduction, the first step of the catalytic cycle. It is shown that the mutation enhances the stability of both enzyme-substrate-complex and the transition state, as well as in ligand binding to the active site when the flavin is in the semiquinone state. The first electron transfer step, from the reduced flavin to heme, is not affected by the mutation, but indeed affects the second electron transfer from flavin semiquinone to heme b(2). | + | The mutation has altered the flavin reduction, the first step of the catalytic cycle. It is shown that the mutation enhances the stability of both enzyme-substrate-complex and the transition state, as well as in ligand binding to the active site when the flavin is in the semiquinone state. The first electron transfer step, from the reduced flavin to heme, is not affected by the mutation, but it indeed affects the second electron transfer from flavin semiquinone to heme b(2). |
The resolution through X-ray crystal structure R289K-b(2) has been determined to 2,75 Å. | The resolution through X-ray crystal structure R289K-b(2) has been determined to 2,75 Å. | ||
Revision as of 22:35, 10 January 2019
| This Sandbox is Reserved from 06/12/2018, through 30/06/2019 for use in the course "Structural Biology" taught by Bruno Kieffer at the University of Strasbourg, ESBS. This reservation includes Sandbox Reserved 1480 through Sandbox Reserved 1543. |
To get started:
More help: Help:Editing |
| |||||||||||
References
- ↑ 1.0 1.1 NYGAARD AP. Various forms of D- and L-lactate dehydrogenases in yeast. Ann N Y Acad Sci. 1961 Nov 2;94:774-9. PMID:14480786
- ↑ doi: https://dx.doi.org/10.1016/0076-6879(66)09064-5
- ↑ Jacq C, Lederer F. [Two molecular species of cytochrome b 2 from Saccharomyces cerevisiae]. Eur J Biochem. 1972 Jan 31;25(1):41-8. PMID:4336855
- ↑ Lederer F. On the first steps of lactate oxidation by bakers' yeast L-(plus)-lactate dehydrogenase (cytochrome b2). Eur J Biochem. 1974 Jul 15;46(2):393-9. PMID:4152980
- ↑ Guiard B, Groudinsky O, Lederer F. Yeast L-lactate dehydrogenase (cytochrome b 2 ). Chemical characterization of the heme-binding core. Eur J Biochem. 1973 Apr;34(2):241-7. PMID:4575975
- ↑ Guiard B, Groudinsky O, Lederer F. Homology between bakers' yeast cytochrome b2 and liver microsomal cytochrome b5. Proc Natl Acad Sci U S A. 1974 Jun;71(6):2539-43. PMID:4210211
- ↑ APPLEBY CA, MORTON RK. Lactic dehydrogenase and cytochrome b2 of baker's yeast; purification and crystallization. Biochem J. 1959 Mar;71(3):492-9. PMID:13638255
- ↑ Lederer F. On the first steps of lactate oxidation by bakers' yeast L-(plus)-lactate dehydrogenase (cytochrome b2). Eur J Biochem. 1974 Jul 15;46(2):393-9. PMID:4152980
- ↑ 9.0 9.1 Tegoni M, Cambillau C. The 2.6-A refined structure of the Escherichia coli recombinant Saccharomyces cerevisiae flavocytochrome b2-sulfite complex. Protein Sci. 1994 Feb;3(2):303-13. PMID:8003966