Sandbox Reserved 1507
From Proteopedia
(Difference between revisions)
| Line 14: | Line 14: | ||
Hsp70 is a family of proteins. They chaperone plenty of cellular proteins and their role is to help the good folding of proteins by preventing aggregation and degradation and help protein transport across membranes. | Hsp70 is a family of proteins. They chaperone plenty of cellular proteins and their role is to help the good folding of proteins by preventing aggregation and degradation and help protein transport across membranes. | ||
| - | Hsp70 is composed of two chains and is 382 nucleotides long. It’s molecular weight is 70 kDa. It can be linked to different ligands such as ATP and ions like potassium or magnesium. | + | Hsp70 is composed of two chains and is 382 nucleotides long. It’s molecular weight is 70 kDa. It can be linked to different ligands such as ATP and ions like potassium or magnesium. <ref>DOI 10.1016/j.cell.2008.05.022</ref> |
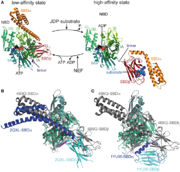
They are composed of a N-terminal ATP binding domain called Nucleotide Binding Domain (NBD) and a C-terminal Peptide-Binding Domain (PDB). The PDB domain is composed of a ß-sandwich part and an α-helical bundle. | They are composed of a N-terminal ATP binding domain called Nucleotide Binding Domain (NBD) and a C-terminal Peptide-Binding Domain (PDB). The PDB domain is composed of a ß-sandwich part and an α-helical bundle. | ||
The aim of the NBD is to regulate the activity of PDB for the substrate binding and release. | The aim of the NBD is to regulate the activity of PDB for the substrate binding and release. | ||
| Line 40: | Line 40: | ||
== Complex activity == | == Complex activity == | ||
| - | The activity of the protein Hsp70 is tightly regulated by different proteins. One kind of proteins will help the hydrolysis of ATP (J-domain proteins) and NEFs (Nucleotide Exchange Factor) which are another kind of proteins will remove the ADP from Hsp70. Sse1 is a NEF, its main function is to catalyse the nucleotide exchange on Hsp70, thereby increasing the rate of the activity of Hsp70 | + | The activity of the protein Hsp70 is tightly regulated by different proteins. One kind of proteins will help the hydrolysis of ATP (J-domain proteins) and NEFs (Nucleotide Exchange Factor) which are another kind of proteins will remove the ADP from Hsp70.<ref> DOI: 10.1016/j.cell.2008.05.022 </ref> Sse1 is a NEF, its main function is to catalyse the nucleotide exchange on Hsp70, thereby increasing the rate of the activity of Hsp70. |
In the activity cycle of Hsp70, unfolded protein are recruited by Hsp70 with the help of J-domain proteins. J-domain proteins binding triggers the ATP hydrolysis in Hsp70 protein. The transformation of ATP into ADP leads to important conformational changes : the PDB domain of Hsp70 adopts a close conformation and binds tightly to the substrate. It’s at this step that intervene Sse1ps. They interact both with Hsp70 and the unfolded protein. The interaction of both the unfolded protein and the chaperone might promote the formation of the complex. | In the activity cycle of Hsp70, unfolded protein are recruited by Hsp70 with the help of J-domain proteins. J-domain proteins binding triggers the ATP hydrolysis in Hsp70 protein. The transformation of ATP into ADP leads to important conformational changes : the PDB domain of Hsp70 adopts a close conformation and binds tightly to the substrate. It’s at this step that intervene Sse1ps. They interact both with Hsp70 and the unfolded protein. The interaction of both the unfolded protein and the chaperone might promote the formation of the complex. | ||
| Line 55: | Line 55: | ||
</StructureSection> | </StructureSection> | ||
== References == | == References == | ||
| - | <references/> | ||
| - | Frontiers in Molecular Biosciences, Insights into the molecular mechanism of allostery in Hsp70s | ||
| - | https://www.frontiersin.org/articles/10.3389/fmolb.2015.00058/full | ||
| - | |||
| - | <references/> | ||
| - | Structural Basis for the Cooperation of Hsp70 and Hsp110 Chaperones in Protein Folding | ||
| - | Sigrun Polier, Zdravko Dragovic, F. Ulrich Harti, Andreas Bracher | ||
| - | Cell 133, 1068-1079, June 13, 2008 | ||
| - | |||
[PubMed Abstract] | [PubMed Abstract] | ||
Ballinger, C. A., Connell, P., Wu, Y., Hu, Z., Thompson, L. J., Yin, L. Y., et al. (1999). Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol. Cell. Biol. 19, 4535–4545. | Ballinger, C. A., Connell, P., Wu, Y., Hu, Z., Thompson, L. J., Yin, L. Y., et al. (1999). Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol. Cell. Biol. 19, 4535–4545. | ||
Revision as of 12:47, 11 January 2019
| This Sandbox is Reserved from 06/12/2018, through 30/06/2019 for use in the course "Structural Biology" taught by Bruno Kieffer at the University of Strasbourg, ESBS. This reservation includes Sandbox Reserved 1480 through Sandbox Reserved 1543. |
To get started:
More help: Help:Editing |
3D2F, Crystal structure of a complex of Sse1p and Hsp70
| |||||||||||
References
[PubMed Abstract] Ballinger, C. A., Connell, P., Wu, Y., Hu, Z., Thompson, L. J., Yin, L. Y., et al. (1999). Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol. Cell. Biol. 19, 4535–4545.