User:Estelle Blochouse/ Sandbox 1497
From Proteopedia
(Difference between revisions)
| Line 8: | Line 8: | ||
== Function == | == Function == | ||
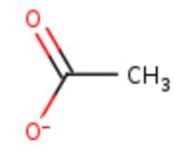
Multicopper oxidases are enzymes involved in copper homeostasis. Copper as many metal ions is used in multiple biological processes such as detoxification of oxygen free radicals and pigmentation. However copper present in a cell bot bounded to a protein if harmfull and can cause cellular damage. It need to be regulated. | Multicopper oxidases are enzymes involved in copper homeostasis. Copper as many metal ions is used in multiple biological processes such as detoxification of oxygen free radicals and pigmentation. However copper present in a cell bot bounded to a protein if harmfull and can cause cellular damage. It need to be regulated. | ||
| - | + | ||
| + | Multicopper oxidase act probably for the detoxification of copper present in the periplasmic space. It oxidize the Cu+ into Cu2+ and prevent its uptake by the cytoplasm. It also possesses a phenoloxidase and ferroxidase activities which can be involved in the prevention of oxidative damage. | ||
| + | |||
Multicopper oxidase might also be involved in the regulation of metal transport. | Multicopper oxidase might also be involved in the regulation of metal transport. | ||
| Line 32: | Line 34: | ||
<tr id='production and extraction'><td class="sblockLbl"><b>Production and extraction:</b></td><td class="sblockDat">Escherichia Coli</td></tr> | <tr id='production and extraction'><td class="sblockLbl"><b>Production and extraction:</b></td><td class="sblockDat">Escherichia Coli</td></tr> | ||
<tr id='initial gene'><td class="sblockLbl"><b>Initial gene:</b></td><td class="sblockDat">sequence from amino acid 29 to amino acid 516 from P36649</td></tr> | <tr id='initial gene'><td class="sblockLbl"><b>Initial gene:</b></td><td class="sblockDat">sequence from amino acid 29 to amino acid 516 from P36649</td></tr> | ||
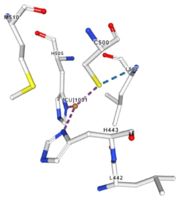
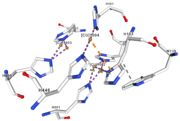
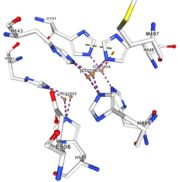
| + | <tr id='specific region'><td class="sblockLbl"><b>Specific region:</b></td><td class="sblockDat">contain a methionine rich region that can be important for copper tolerance in bacteria</td></tr> | ||
<tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=4e9s FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=4e9s OCA], [http://pdbe.org/4e9s PDBe], [http://www.rcsb.org/pdb/explore.do?structureId=4e9s RCSB], [http://www.ebi.ac.uk/pdbsum/4e9s PDBsum], [http://prosat.h-its.org/prosat/prosatexe?pdbcode=4e9s ProSAT], [https://www.ebi.ac.uk/pdbe/entry/pdb/4E9S EMBL-EBI]</span></td></tr> | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=4e9s FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=4e9s OCA], [http://pdbe.org/4e9s PDBe], [http://www.rcsb.org/pdb/explore.do?structureId=4e9s RCSB], [http://www.ebi.ac.uk/pdbsum/4e9s PDBsum], [http://prosat.h-its.org/prosat/prosatexe?pdbcode=4e9s ProSAT], [https://www.ebi.ac.uk/pdbe/entry/pdb/4E9S EMBL-EBI]</span></td></tr> | ||
</table> | </table> | ||
Revision as of 14:36, 11 January 2019
Multicopper Oxidase CueO (4e9s)
| |||||||||||
References
- ↑ EMBL-EBI, Family: Cu-oxidase (PF00394), Summary: Multicopper oxidase, http://pfam.xfam.org/family/Cu-oxidase
- ↑ <ref>PMID:9556453</ref>
- ↑ <ref>PMID:9556453</ref>
- ↑ <ref>PMID:9556453</ref>
- ↑ RCSB PDB
- ↑ RCSB PDB
- ↑ RCSB PDB
- ↑ RCSB PDB
- ↑ RCSB PDB
- ↑ RCSB PDB
- ↑ Kataoka K, Komori H, Ueki Y, Konno Y, Kamitaka Y, Kurose S, Tsujimura S, Higuchi Y, Kano K, Seo D, Sakurai T. Structure and function of the engineered multicopper oxidase CueO from Escherichia coli--deletion of the methionine-rich helical region covering the substrate-binding site. J Mol Biol. 2007 Oct 12;373(1):141-52. Epub 2007 Aug 2. PMID:17804014 doi:10.1016/j.jmb.2007.07.041