Main Page
From Proteopedia
| Line 8: | Line 8: | ||
<span style="border:none; margin:0; padding:0.3em; color:#000; font-style: italic; font-size: 1.1em;max-width:80%;display:block;"> | <span style="border:none; margin:0; padding:0.3em; color:#000; font-style: italic; font-size: 1.1em;max-width:80%;display:block;"> | ||
| - | + | Its goal is to present this information in a user-friendly manner to a broad scientific audience as a free, collaborative 3D-encyclopedia of proteins & other molecules. | |
</span> | </span> | ||
Revision as of 09:05, 20 January 2019
|
ISSN 2310-6301
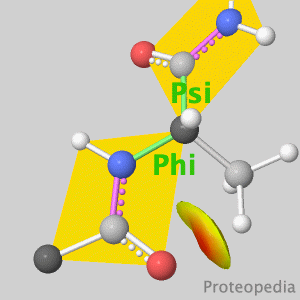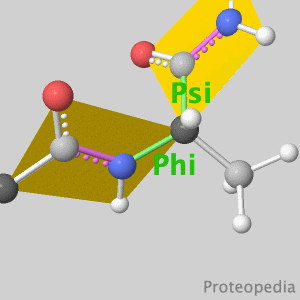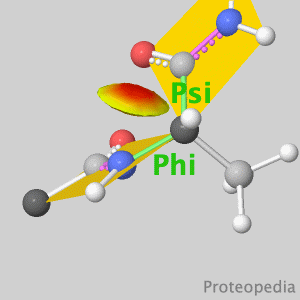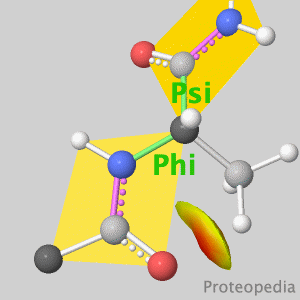
As life is more than 2D, Proteopedia helps to bridge the gap between 3D structure & function of biomacromolecules Its goal is to present this information in a user-friendly manner to a broad scientific audience as a free, collaborative 3D-encyclopedia of proteins & other molecules.
| ||||||||
| Selected Pages | Journals | Education | ||||||
|---|---|---|---|---|---|---|---|---|
|
|
|
||||||
|
How to add content to Proteopedia Who knows ... |
Teaching strategies using Proteopedia |
|||||||
| ||||||||