Journal:Acta Cryst F:S2053230X19001213
From Proteopedia
(Difference between revisions)

| Line 10: | Line 10: | ||
<scene name='80/806392/Cv1/6'>Binding modes of substrate analogues</scene>. The MtDPPS dimer is superimposed on itself with the two polypeptide chains switched. The protein is colored cyan/green in one dimer and pink/yellow in the other, and so are the side chains and the ligands, which are shown as stick models. Mg and water molecules are shown as spheres, and the coordinate bonds as dashed lines. Location of the S1 and S2 site as well as the nearby helices α1/α2 and strand βB are also indicated. When the S1 and S2 substrates and Mg are properly bound for catalysis, the <scene name='80/806392/Cv1/9'>Asp76 side chain not only binds directly to Mg but also to a coordinating water molecule</scene>. The <scene name='80/806392/Cv1/10'>same water is hydrogen bonded to the side chain of Arg292*, which also binds to the other Mg-bound water</scene>. In the absence of both the S2 substrate and Mg, <scene name='80/806392/Cv1/11'>Arg292* turns to bind directly to Asp76</scene>, which is no longer engaged in Mg-coordination. The <scene name='80/806392/Cv1/12'>side chain of Arg292* binds to the β-phosphate of the S1 substrate in this conformation</scene>, and in the other it is also close to the β-phosphate of the S2 substrate. | <scene name='80/806392/Cv1/6'>Binding modes of substrate analogues</scene>. The MtDPPS dimer is superimposed on itself with the two polypeptide chains switched. The protein is colored cyan/green in one dimer and pink/yellow in the other, and so are the side chains and the ligands, which are shown as stick models. Mg and water molecules are shown as spheres, and the coordinate bonds as dashed lines. Location of the S1 and S2 site as well as the nearby helices α1/α2 and strand βB are also indicated. When the S1 and S2 substrates and Mg are properly bound for catalysis, the <scene name='80/806392/Cv1/9'>Asp76 side chain not only binds directly to Mg but also to a coordinating water molecule</scene>. The <scene name='80/806392/Cv1/10'>same water is hydrogen bonded to the side chain of Arg292*, which also binds to the other Mg-bound water</scene>. In the absence of both the S2 substrate and Mg, <scene name='80/806392/Cv1/11'>Arg292* turns to bind directly to Asp76</scene>, which is no longer engaged in Mg-coordination. The <scene name='80/806392/Cv1/12'>side chain of Arg292* binds to the β-phosphate of the S1 substrate in this conformation</scene>, and in the other it is also close to the β-phosphate of the S2 substrate. | ||
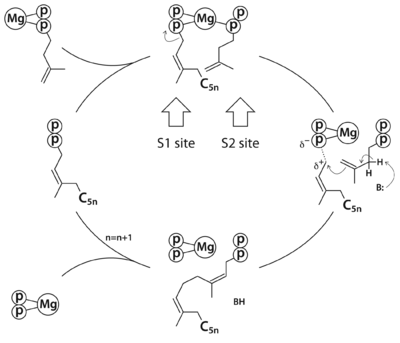
| - | + | The reaction catalyzed by Rv2361c (or ''M. tuberculosis'' DPP synthase, MtDPPS) is very similar to that of undecaprenyl diphosphate synthase (UPPS), except for the chain length of the final product (C<sub>50</sub> vs C<sub>55</sub>) and the starting allylic substrate (''EZ''-FPP vs ''EE''-FPP). In fact, most ''cis''-PTs share a common dimeric architecture, and the conserved S1 and S2 sites for substrate binding are located near the subunit interface. The starting allylic substrate is bound to the S1 site and the homoallylic substrate to be incorporated is bound to the S2 site. An invariant aspartate residue plays a central role in the catalysis by coordinating the Mg2+-bound substrates. The head-to-tail coupling reaction of ''cis''-PT proceeds through a concerted pathway similar to the ionization-condensation-elimination mechanism of ''trans''-PT. After the new C-C bond formation, the pyrophosphate leaves the S1 site along with Mg2+, and the resulting prenyl diphosphate switches from the S2 site to the S1 site (see static image below). | |
[[Image:RxnPathway.png|left|400px|thumb| The reaction pathway of ''cis''-prenyltransferase in general. C<sub>5n</sub> stands for a hydrocarbon group of n prenyl units.]] | [[Image:RxnPathway.png|left|400px|thumb| The reaction pathway of ''cis''-prenyltransferase in general. C<sub>5n</sub> stands for a hydrocarbon group of n prenyl units.]] | ||
Revision as of 09:41, 30 January 2019
| |||||||||||
This page complements a publication in scientific journals and is one of the Proteopedia's Interactive 3D Complement pages. For aditional details please see I3DC.