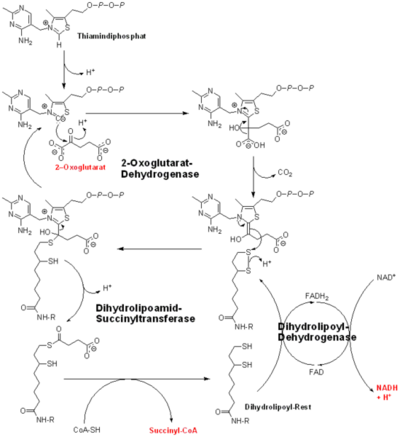
2-Oxoglutarate Dehydrogenase
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
| - | |||
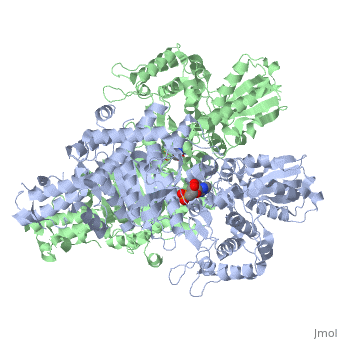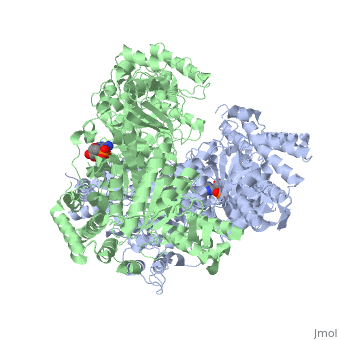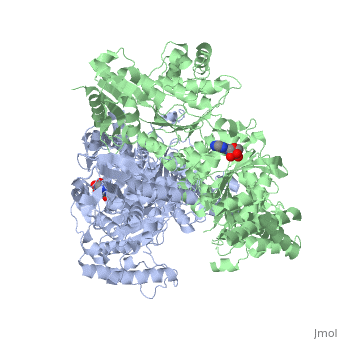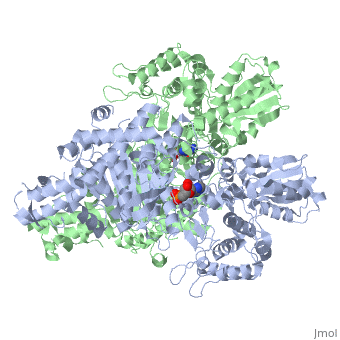
<StructureSection load='2jgd' size='400' side='right' scene='' caption='Dimer of E. coli 2-oxogluterate dehydrogenase E1 component complex with AMP [[2jgd]]'> | <StructureSection load='2jgd' size='400' side='right' scene='' caption='Dimer of E. coli 2-oxogluterate dehydrogenase E1 component complex with AMP [[2jgd]]'> | ||
| Line 23: | Line 22: | ||
==Transfer to E2o== | ==Transfer to E2o== | ||
The overall complex (all of the sub units) help catalysis by keeping the necessary substrates for the reaction close within the enzyme so that it is more likely that the substrate will be in favorable conformation. This enzyme is also part of a larger multienzyme complex that channels the intermediates in the catalysis between subunits of the complex thus minimizing unwanted side reactions<ref>Voet, Donald, Judith G. Voet, and Charlotte W. Pratt. Fundamentals of Biochemistry: Life at the Molecular Level. Hoboken, NJ: Wiley, 2008. p.580</ref>. Not only do the subunits ferry products back and forth but each of the mers in the E1o homodimer are connected via a cavity lined with acidic residues thus increasing the dimer's ability to act as a base. | The overall complex (all of the sub units) help catalysis by keeping the necessary substrates for the reaction close within the enzyme so that it is more likely that the substrate will be in favorable conformation. This enzyme is also part of a larger multienzyme complex that channels the intermediates in the catalysis between subunits of the complex thus minimizing unwanted side reactions<ref>Voet, Donald, Judith G. Voet, and Charlotte W. Pratt. Fundamentals of Biochemistry: Life at the Molecular Level. Hoboken, NJ: Wiley, 2008. p.580</ref>. Not only do the subunits ferry products back and forth but each of the mers in the E1o homodimer are connected via a cavity lined with acidic residues thus increasing the dimer's ability to act as a base. | ||
| + | |||
| + | ==3D structues of 2-oxoglutarate dehydrogenase== | ||
| + | [[2-oxoglutarate dehydrogenase 3D structures]] | ||
</StructureSection> | </StructureSection> | ||
Revision as of 09:15, 20 February 2019
| |||||||||||
3D structures of 2-Oxoglutarate dehydrogenase
Updated on 20-February-2019
References
- ↑ 1.0 1.1 1.2 1.3 Frank RA, Price AJ, Northrop FD, Perham RN, Luisi BF. Crystal structure of the E1 component of the Escherichia coli 2-oxoglutarate dehydrogenase multienzyme complex. J Mol Biol. 2007 May 4;368(3):639-51. Epub 2007 Feb 7. PMID:17367808 doi:10.1016/j.jmb.2007.01.080
- ↑ 2.0 2.1 McMinn CL, Ottaway JH. Studies on the mechanism and kinetics of the 2-oxoglutarate dehydrogenase system from pig heart. Biochem J. 1977 Mar 1;161(3):569-81. PMID:192200
- ↑ Leung PS, Rossaro L, Davis PA, Park O, Tanaka A, Kikuchi K, Miyakawa H, Norman GL, Lee W, Gershwin ME. Antimitochondrial antibodies in acute liver failure: implications for primary biliary cirrhosis. Hepatology. 2007 Nov;46(5):1436-42. PMID:17657817 doi:10.1002/hep.21828
- ↑ Voet, Donald, Judith G. Voet, and Charlotte W. Pratt. Fundamentals of Biochemistry: Life at the Molecular Level. Hoboken, NJ: Wiley, 2008. p.580
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, David Canner, Lucas Evans, Alexander Berchansky, Joel L. Sussman