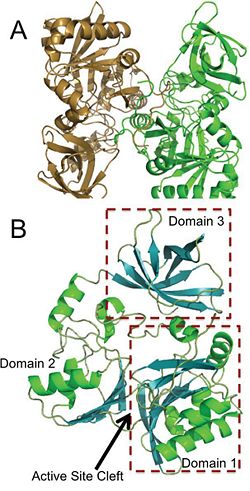
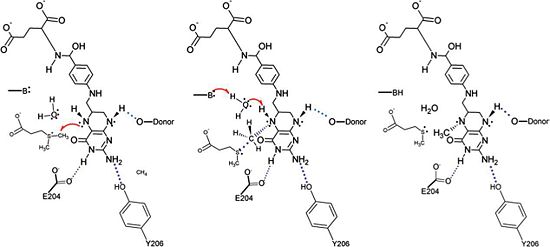
Aminomethyltransferase
From Proteopedia
(Difference between revisions)
| Line 27: | Line 27: | ||
DMSP and its degradation products, particularly dimethyl sulfide (DMS), play a key role in the sulfur cycle. Therefore a complete understanding of both the cleavage and demethylation pathways as well as the enzymes involved could have significant environmental implications. In particular, it currently appears that certain types of organisms such as ''Pelagabacter ubique,'' the microbe that DmdA was originally isolated from, utilize the demethylation pathway over the cleavage pathway because it provides an additional carbon energy source. The ability to not only identify the organisms that degrade DMSP, but also the conditions under which each pathway is more favorable, could have significant biogeochemical and bioremediation applications. However, there is a significant amount of research that must be conducted before those applications can even begin to be considered. Current research is focused on identifying and characterizing DMSP-degrading organisms as well as the role of DmdA in certain environmental conditions, such an induced phytoplankton bloom<ref>Howard, E.C., Sun, S., Reisch, C.R., del Valle, D.A>, Burgmann, H., Kiene, R.P., and Moran, M.A. (2011). Changes in dimethylsuloniopropionate demethylase gene assemblages in response to an induced phytoplankton bloom. Appl Environ Microbiol. 77(2):524-531.</ref>. For example, oceanic metagenomic data has been collected and analyzed in order to determine what marine microorganisms contain the gene to produce DmdA <ref>Howard, E.C., Sun, S., Biers, E.J., and Moran, M.A. (2008). Abundant and diverse bacteria involved in DMSP degradation in marine surface waters. Environ Microbiol. 10(9);2397-2410.</ref>. The results of these analysis suggest that a significant percentage of marine bacteria are involved in DMSP demethylation, as the gene to produce DmdA has been found among a wide range of diverse taxa. | DMSP and its degradation products, particularly dimethyl sulfide (DMS), play a key role in the sulfur cycle. Therefore a complete understanding of both the cleavage and demethylation pathways as well as the enzymes involved could have significant environmental implications. In particular, it currently appears that certain types of organisms such as ''Pelagabacter ubique,'' the microbe that DmdA was originally isolated from, utilize the demethylation pathway over the cleavage pathway because it provides an additional carbon energy source. The ability to not only identify the organisms that degrade DMSP, but also the conditions under which each pathway is more favorable, could have significant biogeochemical and bioremediation applications. However, there is a significant amount of research that must be conducted before those applications can even begin to be considered. Current research is focused on identifying and characterizing DMSP-degrading organisms as well as the role of DmdA in certain environmental conditions, such an induced phytoplankton bloom<ref>Howard, E.C., Sun, S., Reisch, C.R., del Valle, D.A>, Burgmann, H., Kiene, R.P., and Moran, M.A. (2011). Changes in dimethylsuloniopropionate demethylase gene assemblages in response to an induced phytoplankton bloom. Appl Environ Microbiol. 77(2):524-531.</ref>. For example, oceanic metagenomic data has been collected and analyzed in order to determine what marine microorganisms contain the gene to produce DmdA <ref>Howard, E.C., Sun, S., Biers, E.J., and Moran, M.A. (2008). Abundant and diverse bacteria involved in DMSP degradation in marine surface waters. Environ Microbiol. 10(9);2397-2410.</ref>. The results of these analysis suggest that a significant percentage of marine bacteria are involved in DMSP demethylation, as the gene to produce DmdA has been found among a wide range of diverse taxa. | ||
| + | |||
| + | =3D structure of aminomethyltransferase= | ||
| + | [[Aminomethyltransferase 3D structures]] | ||
| + | |||
</StructureSection> | </StructureSection> | ||
==3D structure of aminomethyltransferase== | ==3D structure of aminomethyltransferase== | ||
Revision as of 09:52, 6 March 2019
| |||||||||||
3D structure of aminomethyltransferase
Updated on 06-March-2019
1woo, 1wop, 1wor, 1wos – AMT – Thermotoga maritima
1vlo – EcAMT (mutant) – Escherichia coli
1v5v – AMT – Pyrococcus horikoshii
1yx2 – AMT – Bacillus subtilis
1wsr, 1wsv – AMT – human
3gir – AMT – Bartonella henselae
3tfh – CpAMT-like – Candidatus pelagibacter
3tfi – CpAMT-like + DMSP
3tfj – CpAMT-like + tetrahydrofolate
3a8i, 3a8j – EcAMT + glycine cleavage system H protein
3a8k – EcAMT (mutant) + glycine cleavage system H protein
References
- ↑ Reisch, C.R., Moran, M.A., Whitman, W.B. (2008). Dimethylsulfoniopropionate-Dependent Demethylase (DmdA) from Pelagibacter ubique and Silicibacter pomeroyi. J. Bacteriol. 190: 8018-8024.
- ↑ Malin, G. (2006). New Pieces for the Marine Sulfur Cycle Jigsaw. Science. 314: 607-608.
- ↑ Image from the RCSB PDB (www.pdb.org) of PDB ID 3TFH (Schuller, D.J., Reisch, C.R., Moran, M.A., Whitman, W.B., Lanzilotta, W.N. (2012) Structures of dimethylsulfoniopropinate-dependent demethylase from the marine organism pelagabacter ubique. Protein Sci. 21: 289-298).
- ↑ Schuller, D.J., Reisch, C.R., Moran, M.A., Whitman, W.B., Lanzilotta, W.N. (2012) Structures of dimethylsulfoniopropinate-dependent demethylase from the marine organism pelagabacter ubique. Protein Sci. 21: 289-298.
- ↑ Howard, E.C., Sun, S., Reisch, C.R., del Valle, D.A>, Burgmann, H., Kiene, R.P., and Moran, M.A. (2011). Changes in dimethylsuloniopropionate demethylase gene assemblages in response to an induced phytoplankton bloom. Appl Environ Microbiol. 77(2):524-531.
- ↑ Howard, E.C., Sun, S., Biers, E.J., and Moran, M.A. (2008). Abundant and diverse bacteria involved in DMSP degradation in marine surface waters. Environ Microbiol. 10(9);2397-2410.
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Joel L. Sussman, Kara Tinker, Jaime Prilusky