User:Jordan Finch/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 13: | Line 13: | ||
== Mechanism == | == Mechanism == | ||
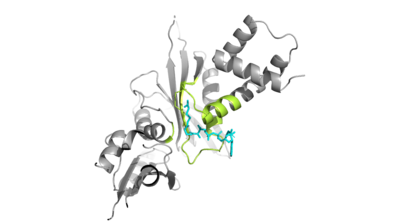
| - | Of the | + | Of the five classes of HAT enzymes, the catalytic mechanism for HAT1 and Rtt109 remains unclear. A structural overlay of HAT1 and Gcn5, a more well-known and understood HAT enzyme, found a conserved glutamate residue in the active site of both molecules. It was found that mutation at that active site glutamate residue greatly alters the catalytic ability of HAT1 and has been proven to be structurally important. <ref> DOI:10.1101/gad.240531.114 </ref> Using this information and structurl information regarding the proximity of potentially catalytic residues, the most plausible mechanism for histone acetylation involves the following relevant residues and cofactors <scene name='81/811713/Mechanism_glu_lys_coa/1'>Glu255, Lys14, and AcetylCoA</scene>. |
== Application == | == Application == | ||
== References == | == References == | ||
| - | Li, Y. et. al. ''Hat2p recognizes the histone H3 tail to specify the acetylation of the newly synthesized H3/H4 heterodimer by the Hat1p/Hat2p complex.''(2014). ''Genes Dev.''28:1217-1227. DOI:10.1101/gad.240531.114 | + | 1. Li, Y. et. al. ''Hat2p recognizes the histone H3 tail to specify the acetylation of the newly synthesized H3/H4 heterodimer by the Hat1p/Hat2p complex.''(2014). ''Genes Dev.''28:1217-1227. DOI:10.1101/gad.240531.114 |
<references/> | <references/> | ||
Revision as of 21:56, 4 April 2019
Histone Acetyltransferase HAT1/HAT2 Complex, Saccharomyces cerevisiae
| |||||||||||