This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
User:Jordan Finch/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 13: | Line 13: | ||
== Mechanism == | == Mechanism == | ||
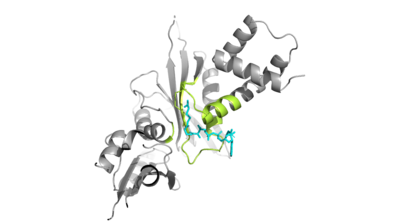
| - | Of the five classes of HAT enzymes, the catalytic mechanisms for two of those enzymes, HAT1 and Rtt109, remains unclear. A structural overlay of HAT1 and Gcn5, a more well-known and understood HAT enzyme, found a conserved glutamate residue in the active site of both molecules. It was found that mutation at that active site glutamate residue greatly alters the catalytic ability of HAT1 and has been proven to be structurally important. <ref> DOI:10.1101/gad.240531.114 </ref> Using this information and | + | Of the five classes of HAT enzymes, the catalytic mechanisms for two of those enzymes, HAT1 and Rtt109, remains unclear. A structural overlay of HAT1 and Gcn5, a more well-known and understood HAT enzyme, found a conserved glutamate residue in the active site of both molecules. It was found that mutation at that active site glutamate residue greatly alters the catalytic ability of HAT1 and has been proven to be structurally important. <ref> DOI:10.1101/gad.240531.114 </ref> Using this information and structural information regarding the proximity of potentially catalytic residues, the most plausible mechanism for histone acetylation involves the following relevant residues and cofactors <scene name='81/811713/Mechanism_glu_lys_coa/1'>Glu255, Lys14, and AcetylCoA</scene>. |
| + | |||
| + | In this mechanism, the glutamate at residue 255 acts as a general base and deprotonates lysine 12 of histone 4 (the numbering of the modified lysine residue on histone 4 is shifted two residues).The deprotonated lysine acts as a nucleophile and attacts the carbonyl carbon of acetyl coenzyme A | ||
== Application == | == Application == | ||
Revision as of 17:32, 5 April 2019
Histone Acetyltransferase HAT1/HAT2 Complex, Saccharomyces cerevisiae
| |||||||||||