Introduction
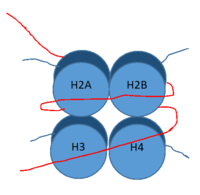
DNA (red) wrapped around histone proteins with histone tails (blue)
LSD-1 is a lysine demethylase. A histone is a blah blah blah and can be seen in Fig. 1.
Structure

Figure 1:LSD1 overall 3D structure: Tower domain (blue), SWIRM domain (yellow), and Oxidase domain (red).
Tower Domain
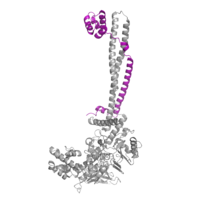
Figure 2: CoRest complex (purple) bound to LSD1 at the Tower domain.
The is a protrusion off the main protein body of LSD-1 comprised of 100 residues, which form 2 𝛂-helices. The longer helix, T𝛂A, is an LSD-1 specific element that has not been found in any other oxidase proteins [1]. The shorter helix, T𝛂B, is very near to the active site of the oxidase domain. In fact, T𝛂B connects directly to helix 𝛂D of the oxidase domain through a highly conserved connector loop. The exact function of the tower domain is not known, but it is proposed to regulate the size of the active site chamber through this . The T𝛂B-𝛂D interaction is responsible for the proper positioning of , a side chain of 𝛂D that is located in the catalytic chamber. In addition, the T𝛂B-𝛂D interaction positions 𝛂D in the correct manner to provide hydrogen bonding to . Tyr761 is positioned in the catalytic chamber very close to the FAD cofactor, and aids in the binding of the lysine substrate [1]. Therefore, the base of the tower domain forms a direct connection to the oxidase domain and plays a crucial role in the shape and catalytic activity of the active site. In fact, removing the tower domain via a mutation resulted in a drastic decrease in catalytic efficiency [1]. The tower domain has also been found to interact with other proteins and complexes, such as CoREST (Figure 2), as a molecular lever to allosterically regulate the catalytic activity of the active site [2]. Overall, the exact function of the tower domain has not yet been determined, but it is known to be vital to the catalytic activity of LSD-1.
SWIRM Domain
Oxidase Domain
Active Site and FAD Cofactor
Mechanism of Active Site

Figure X: Hydride transfer mechanism of LSD-1 active site via FAD cofactor.
This is a picture of the .
These are the different
The is ALSO a blah blah blah.
Inhibition by Tri-Methylated Lysine
Medical Implications
1. Cancer
2. Diabetes
References
- ↑ 1.0 1.1 1.2 Stavropoulos P, Blobel G, Hoelz A. Crystal structure and mechanism of human lysine-specific demethylase-1. Nat Struct Mol Biol. 2006 Jul;13(7):626-32. Epub 2006 Jun 25. PMID:16799558 doi:10.1038/nsmb1113
- ↑ Yang M, Gocke CB, Luo X, Borek D, Tomchick DR, Machius M, Otwinowski Z, Yu H. Structural basis for CoREST-dependent demethylation of nucleosomes by the human LSD1 histone demethylase. Mol Cell. 2006 Aug 4;23(3):377-87. PMID:16885027 doi:http://dx.doi.org/10.1016/j.molcel.2006.07.012



