We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Nicholas Bantz/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 7: | Line 7: | ||
== Structure == | == Structure == | ||
| - | [[Image:LSD1 Pic.png|200 px|left|thumb|Figure 2: LSD1 overall 3D structure: Tower domain (blue), SWIRM domain (yellow), and Oxidase domain (red), FAD cofactor (green).]] | ||
=== Tower Domain === | === Tower Domain === | ||
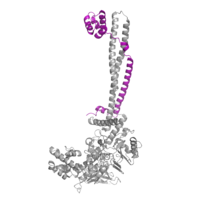
| - | [[Image:COREST.png|200 px|right|thumb|Figure | + | [[Image:COREST.png|200 px|right|thumb|Figure 2: CoRest complex (purple) bound to LSD1 at the Tower domain.]] |
| - | The <scene name='81/811088/Towerdomain/2'>tower domain</scene> is a 100 residue protrusion off of the main protein body of LSD-1, comprised of 2 [https://en.wikipedia.org/wiki/Alpha_helix-helices 𝛂-helices]. The longer helix, T𝛂A, is an LSD-1 specific element that has not been found in any other oxidase proteins <ref name="Stavropolous">doi: 10.1038/nsmb1113</ref>. The shorter helix, T𝛂B, is very near to the active site of the oxidase domain. In fact, T𝛂B connects directly to helix 𝛂D of the oxidase domain through a highly conserved connector loop. The exact function of the tower domain is not known, but it is proposed to regulate the size of the active site chamber through this <scene name='81/811090/Tb-dinteraction/1'>TαB-αD interaction</scene>. The T𝛂B-𝛂D interaction is responsible for the proper positioning of <scene name='81/811090/Phe538-tyr761interaction/1'>Phe538</scene>, a side chain of 𝛂D that is located in the catalytic chamber for proper recognition and binding of the substrate lysine through hydrophobic interactions. In addition, the T𝛂B-𝛂D interaction positions 𝛂D in the correct manner to provide [https://en.wikipedia.org/wiki/Hydrogen_bond hydrogen bonding] to <scene name='81/811090/Phe538-tyr761interaction/1'>Tyr761</scene>. Tyr761 is positioned in the catalytic chamber very close to the FAD cofactor, and aids in the binding of the lysine substrate <ref name="Stavropolous"/>. Therefore, the base of the tower domain forms a direct connection to the oxidase domain and plays a crucial role in the shape and catalytic activity of the active site. In fact, removing the tower domain via a mutation resulted in a drastic decrease in catalytic efficiency <ref name="Stavropolous"/>. The tower domain has also been found to interact with other proteins and complexes, such as CoREST (Figure | + | The <scene name='81/811088/Towerdomain/2'>tower domain</scene> is a 100 residue protrusion off of the main protein body of LSD-1, comprised of 2 [https://en.wikipedia.org/wiki/Alpha_helix-helices 𝛂-helices]. The longer helix, T𝛂A, is an LSD-1 specific element that has not been found in any other oxidase proteins <ref name="Stavropolous">doi: 10.1038/nsmb1113</ref>. The shorter helix, T𝛂B, is very near to the active site of the oxidase domain. In fact, T𝛂B connects directly to helix 𝛂D of the oxidase domain through a highly conserved connector loop. The exact function of the tower domain is not known, but it is proposed to regulate the size of the active site chamber through this <scene name='81/811090/Tb-dinteraction/1'>TαB-αD interaction</scene>. The T𝛂B-𝛂D interaction is responsible for the proper positioning of <scene name='81/811090/Phe538-tyr761interaction/1'>Phe538</scene>, a side chain of 𝛂D that is located in the catalytic chamber for proper recognition and binding of the substrate lysine through hydrophobic interactions. In addition, the T𝛂B-𝛂D interaction positions 𝛂D in the correct manner to provide [https://en.wikipedia.org/wiki/Hydrogen_bond hydrogen bonding] to <scene name='81/811090/Phe538-tyr761interaction/1'>Tyr761</scene>. Tyr761 is positioned in the catalytic chamber very close to the FAD cofactor, and aids in the binding of the lysine substrate <ref name="Stavropolous"/>. Therefore, the base of the tower domain forms a direct connection to the oxidase domain and plays a crucial role in the shape and catalytic activity of the active site. In fact, removing the tower domain via a mutation resulted in a drastic decrease in catalytic efficiency <ref name="Stavropolous"/>. The tower domain has also been found to interact with other proteins and complexes, such as CoREST (Figure 2), as a molecular lever to allosterically regulate the catalytic activity of the active site <ref name="Yang">doi: 10.1016/j.molcel.2006.07.012</ref>. Overall, the exact function of the tower domain has not yet been fully determined, but it is known to be vital to the catalytic activity of LSD-1. |
| Line 27: | Line 26: | ||
== Mechanism of Action== | == Mechanism of Action== | ||
| - | [[Image:FADMechanism.png|500 px|right|thumb|Figure | + | [[Image:FADMechanism.png|500 px|right|thumb|Figure 3: Hydride transfer mechanism of LSD-1 active site via FAD cofactor.]] |
| - | The mechanism of lysine demethylation is highly dependent on the presence of the <scene name='81/811089/Fadcofactor/4'>FAD cofactor</scene>. The FAD cofactor, positioned closely to the substrate lysine in the active site, acts as an oxidizing agent and initiates catalysis (Figure | + | The mechanism of lysine demethylation is highly dependent on the presence of the <scene name='81/811089/Fadcofactor/4'>FAD cofactor</scene>. The FAD cofactor, positioned closely to the substrate lysine in the active site, acts as an oxidizing agent and initiates catalysis (Figure 3). A two-electron transfer occurs between the substrate lysine and FAD in the form of a [https://en.wikipedia.org/wiki/Hydride hydride]; the lysine is oxidized and the FAD is reduced <ref name="Stavropolous"/>. The FAD cofactor forms an anion and is stabilized by the positively charged <scene name='81/811088/Lys661/2'>Lys661</scene> positioned in the catalytic pocket of the active site <ref name="Stavropolous"/>. Although Lys661 is 8 Å away from the nitrogen in FAD that is thought to sustain the negative charge in its reduced form, through resonance it is possible that the negative charge may be dispersed to an atom closer to the stabilizing Lys661. The oxidized lysine forms an aminium cation that is hydrolyzed into the carbinolamine intermediate <ref name="Stavropolous"/>. The carbinolamine intermediate readily decomposes into formaldehyde and the demethylated lysine substrate <ref name="Stavropolous"/>. |
===Inhibition by Tri-Methylated Lysine=== | ===Inhibition by Tri-Methylated Lysine=== | ||
| - | [[Image:Tri-methylated Lysine.png|60 px|right|thumb|Figure | + | [[Image:Tri-methylated Lysine.png|60 px|right|thumb|Figure 4: Structure of Tri-methylated lysine; chemical inhibitor of LSD-1 activity.]] |
| - | The proposed LSD-1 mechanism is supported by the fact that tri-methylated lysine substrates (Figure | + | The proposed LSD-1 mechanism is supported by the fact that tri-methylated lysine substrates (Figure 4) competitively inhibit the enzyme. A substrate lysine that is tri-methylated binds to the active site but does not undergo catalysis; the inhibition is not steric (the active site is large enough to accommodate tri-methylated lysines), but is rather chemical in nature. Tri-methylated lysines do not have a free hydrogen to lose in a hydride transfer as is necessitated by the proposed mechanism, resulting in chemical inhibition of LSD-1 <ref name="Stavropolous"/>. Thus, the mechanism of LSD-1 contributes to its specificity for mono- or di-methylated lysine substrates (Figure 4). |
== Medical Implications == | == Medical Implications == | ||
Revision as of 01:59, 9 April 2019
LSD-1: Human lysine-specific demethylase 1
| |||||||||||
Student Contributors
- Nicholas Bantz
- Cody Carley
- Michael Thomas