We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Nicholas Bantz/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 9: | Line 9: | ||
=== Tower Domain === | === Tower Domain === | ||
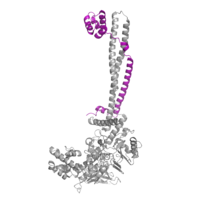
| - | [[Image:COREST.png|200 px| | + | [[Image:COREST.png|200 px|left|thumb|Figure 2: CoRest complex (purple) bound to LSD1 at the Tower domain.]] |
| - | + | ||
The <scene name='81/811088/Towerdomain/2'>tower domain</scene> is a 100 residue protrusion off of the main protein body of LSD-1, comprised of 2 [https://en.wikipedia.org/wiki/Alpha_helix-helices 𝛂-helices]. The longer helix, T𝛂A, is an LSD-1 specific element that has not been found in any other oxidase proteins <ref name="Stavropolous">doi: 10.1038/nsmb1113</ref>. The shorter helix, T𝛂B, is very near to the active site of the oxidase domain. In fact, T𝛂B connects directly to helix 𝛂D of the oxidase domain through a highly conserved connector loop. The exact function of the tower domain is not known, but it is proposed to regulate the size of the active site chamber through this <scene name='81/811090/Tb-dinteraction/1'>TαB-αD interaction</scene>. The T𝛂B-𝛂D interaction is responsible for the proper positioning of <scene name='81/811090/Phe538-tyr761interaction/1'>Phe538</scene>, a side chain of 𝛂D that is located in the catalytic chamber for proper recognition and binding of the substrate lysine through hydrophobic interactions. In addition, the T𝛂B-𝛂D interaction positions 𝛂D in the correct manner to provide [https://en.wikipedia.org/wiki/Hydrogen_bond hydrogen bonding] to <scene name='81/811090/Phe538-tyr761interaction/1'>Tyr761</scene>. Tyr761 is positioned in the catalytic chamber very close to the FAD cofactor, and aids in the binding of the lysine substrate <ref name="Stavropolous"/>. Therefore, the base of the tower domain forms a direct connection to the oxidase domain and plays a crucial role in the shape and catalytic activity of the active site. In fact, removing the tower domain via a mutation resulted in a drastic decrease in catalytic efficiency <ref name="Stavropolous"/>. The tower domain has also been found to interact with other proteins and complexes, such as CoREST (Figure 2), as a molecular lever to allosterically regulate the catalytic activity of the active site <ref name="Yang">doi: 10.1016/j.molcel.2006.07.012</ref>. Overall, the exact function of the tower domain has not yet been fully determined, but it is known to be vital to the catalytic activity of LSD-1. | The <scene name='81/811088/Towerdomain/2'>tower domain</scene> is a 100 residue protrusion off of the main protein body of LSD-1, comprised of 2 [https://en.wikipedia.org/wiki/Alpha_helix-helices 𝛂-helices]. The longer helix, T𝛂A, is an LSD-1 specific element that has not been found in any other oxidase proteins <ref name="Stavropolous">doi: 10.1038/nsmb1113</ref>. The shorter helix, T𝛂B, is very near to the active site of the oxidase domain. In fact, T𝛂B connects directly to helix 𝛂D of the oxidase domain through a highly conserved connector loop. The exact function of the tower domain is not known, but it is proposed to regulate the size of the active site chamber through this <scene name='81/811090/Tb-dinteraction/1'>TαB-αD interaction</scene>. The T𝛂B-𝛂D interaction is responsible for the proper positioning of <scene name='81/811090/Phe538-tyr761interaction/1'>Phe538</scene>, a side chain of 𝛂D that is located in the catalytic chamber for proper recognition and binding of the substrate lysine through hydrophobic interactions. In addition, the T𝛂B-𝛂D interaction positions 𝛂D in the correct manner to provide [https://en.wikipedia.org/wiki/Hydrogen_bond hydrogen bonding] to <scene name='81/811090/Phe538-tyr761interaction/1'>Tyr761</scene>. Tyr761 is positioned in the catalytic chamber very close to the FAD cofactor, and aids in the binding of the lysine substrate <ref name="Stavropolous"/>. Therefore, the base of the tower domain forms a direct connection to the oxidase domain and plays a crucial role in the shape and catalytic activity of the active site. In fact, removing the tower domain via a mutation resulted in a drastic decrease in catalytic efficiency <ref name="Stavropolous"/>. The tower domain has also been found to interact with other proteins and complexes, such as CoREST (Figure 2), as a molecular lever to allosterically regulate the catalytic activity of the active site <ref name="Yang">doi: 10.1016/j.molcel.2006.07.012</ref>. Overall, the exact function of the tower domain has not yet been fully determined, but it is known to be vital to the catalytic activity of LSD-1. | ||
=== SWIRM Domain === | === SWIRM Domain === | ||
| - | |||
The <scene name='81/811088/Swirmdomain/4'>SWIRM domain</scene> is seen in numerous enzymes that participate in histone binding and chromatin modification. The SWIRM domain of LSD-1 is 94 residues long and is comprised of an alpha-helix bundle <ref name="Stavropolous"/>. The longest helix, 𝛂C, separates the two other helix-turn-helix motifs <scene name='81/811088/Swirmmotifs/2'>𝛂A/B and 𝛂D/E </scene> <ref name="Stavropolous"/>. The SWIRM domain is attached to the oxidase domain via hydrophobic [https://en.wikipedia.org/wiki/Van_der_Waals_force van der Waals interactions] between 3 𝛂-helices each domain: <scene name='81/811088/Oxidaseandswirmchillin/4'>𝛂A, 𝛂B, and 𝛂E motifs in the SWIRM domain and 𝛂A, 𝛂B, 𝛂M, motifs in the oxidase domain</scene>. The residues that create this hydrophobic interface (which spans nearly 1680 Ų) are practically invariant across histone-modifying proteins <ref name="Stavropolous"/>. The <scene name='81/811090/Hydrophobic_interface/2'> hydrophobic interface between the oxidase and SWIRM domains</scene> creates a cleft or tunnel that is also present in other chromatin modifying enzymes. This cleft is responsible for binding to DNA in the other enzymes through the presence of positively charged residues in the cleft <ref name="Stavropolous"/>. The SWIRM domain in LSD-1 is unique because the cleft that is formed by the hydrophobic SWIRM-oxidase interactions lacks the positively charged residues common in other enzymes <ref name="Stavropolous"/>. For this reason, it is proposed that the SWIRM cleft is used for binding of a histone tail (on the same histone as the substrate lysine) in order to hold the histone in place. Multiple experiments showed that mutations in hydrophobic residues that form the SWIRM-oxidase interface greatly reduced the catalytic activity of LSD-1 <ref name="Stavropolous"/>. This, and the proximity to the active site in the oxidase domain, exhibit the importance of the SWIRM cleft in the mechanism of LSD-1. | The <scene name='81/811088/Swirmdomain/4'>SWIRM domain</scene> is seen in numerous enzymes that participate in histone binding and chromatin modification. The SWIRM domain of LSD-1 is 94 residues long and is comprised of an alpha-helix bundle <ref name="Stavropolous"/>. The longest helix, 𝛂C, separates the two other helix-turn-helix motifs <scene name='81/811088/Swirmmotifs/2'>𝛂A/B and 𝛂D/E </scene> <ref name="Stavropolous"/>. The SWIRM domain is attached to the oxidase domain via hydrophobic [https://en.wikipedia.org/wiki/Van_der_Waals_force van der Waals interactions] between 3 𝛂-helices each domain: <scene name='81/811088/Oxidaseandswirmchillin/4'>𝛂A, 𝛂B, and 𝛂E motifs in the SWIRM domain and 𝛂A, 𝛂B, 𝛂M, motifs in the oxidase domain</scene>. The residues that create this hydrophobic interface (which spans nearly 1680 Ų) are practically invariant across histone-modifying proteins <ref name="Stavropolous"/>. The <scene name='81/811090/Hydrophobic_interface/2'> hydrophobic interface between the oxidase and SWIRM domains</scene> creates a cleft or tunnel that is also present in other chromatin modifying enzymes. This cleft is responsible for binding to DNA in the other enzymes through the presence of positively charged residues in the cleft <ref name="Stavropolous"/>. The SWIRM domain in LSD-1 is unique because the cleft that is formed by the hydrophobic SWIRM-oxidase interactions lacks the positively charged residues common in other enzymes <ref name="Stavropolous"/>. For this reason, it is proposed that the SWIRM cleft is used for binding of a histone tail (on the same histone as the substrate lysine) in order to hold the histone in place. Multiple experiments showed that mutations in hydrophobic residues that form the SWIRM-oxidase interface greatly reduced the catalytic activity of LSD-1 <ref name="Stavropolous"/>. This, and the proximity to the active site in the oxidase domain, exhibit the importance of the SWIRM cleft in the mechanism of LSD-1. | ||
Revision as of 02:02, 9 April 2019
LSD-1: Human lysine-specific demethylase 1
| |||||||||||
Student Contributors
- Nicholas Bantz
- Cody Carley
- Michael Thomas