We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Steve Klimcak/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 17: | Line 17: | ||
===Hydrophobic Pocket=== | ===Hydrophobic Pocket=== | ||
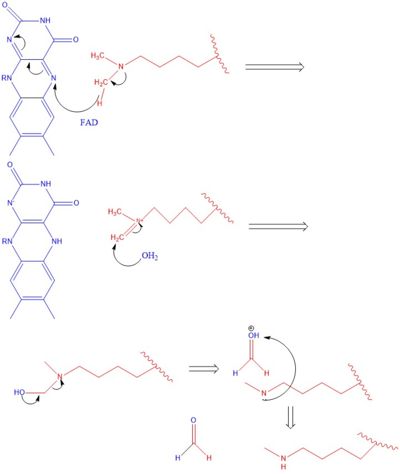
The hydrophobic pocket located in the active site cavity of LSD1, forms a catalytic chamber where the substrate lysine is oriented and positioned to interact with the | The hydrophobic pocket located in the active site cavity of LSD1, forms a catalytic chamber where the substrate lysine is oriented and positioned to interact with the | ||
| - | FAD co-factor to initiate demethylation. The specific residues making up the pocket include valine-317, glycine-330, alanine-331, methionine 332, valine-333, phenylalanine-338, leucine-569, asparagine-660, lysine-661, tryptophan 695, serine 749, serine 760, and tyrosine-761. | + | FAD co-factor to initiate demethylation. The specific residues making up the pocket include valine-317, glycine-330, alanine-331, methionine 332, valine-333, phenylalanine-338, leucine-569, asparagine-660, lysine-661, tryptophan 695, serine 749, serine 760, and tyrosine-761. The <scene name='81/811712/Hydrophobic_pocket/1'>hydrophobic pocket</scene> shown in green surrounds the |
== Disease == | == Disease == | ||
Revision as of 16:27, 9 April 2019
LSD Demethylase
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ Ransey E, Paredes E, Dey SK, Das SR, Heroux A, Macbeth MR. Crystal structure of the Entamoeba histolytica RNA lariat debranching enzyme EhDbr1 reveals a catalytic Zn(2+) /Mn(2+) heterobinucleation. FEBS Lett. 2017 Jul;591(13):2003-2010. doi: 10.1002/1873-3468.12677. Epub 2017, Jun 14. PMID:28504306 doi:http://dx.doi.org/10.1002/1873-3468.12677
- ↑ Khalid MF, Damha MJ, Shuman S, Schwer B. Structure-function analysis of yeast RNA debranching enzyme (Dbr1), a manganese-dependent phosphodiesterase. Nucleic Acids Res. 2005 Nov 7;33(19):6349-60. doi: 10.1093/nar/gki934. Print, 2005. PMID:16275784 doi:http://dx.doi.org/10.1093/nar/gki934