User:Asif Hossain/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 20: | Line 20: | ||
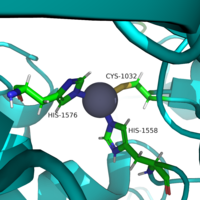
The <scene name='81/811085/Active_site/6'>active site</scene> of HDAC8 is composed of 2 catalytic dyads: <scene name='81/811085/Dyads/2'>His143/Asp183 and His142/Asp176</scene>, which activate the catalytic water nucleophile (Figure 2). A Tyr306, through mutation to Phe in the pdb file 2v5w (modeled in the overall view) was observed to render the protein mostly inactive. Thus, it has been hypothesized that the this residue is critical for stabilization of the transition state with the Zn<sup>2+</sup> ion. | The <scene name='81/811085/Active_site/6'>active site</scene> of HDAC8 is composed of 2 catalytic dyads: <scene name='81/811085/Dyads/2'>His143/Asp183 and His142/Asp176</scene>, which activate the catalytic water nucleophile (Figure 2). A Tyr306, through mutation to Phe in the pdb file 2v5w (modeled in the overall view) was observed to render the protein mostly inactive. Thus, it has been hypothesized that the this residue is critical for stabilization of the transition state with the Zn<sup>2+</sup> ion. | ||
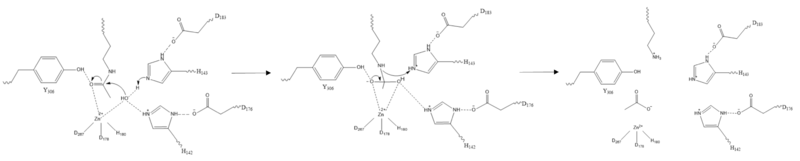
| - | [[Image:Triad.png |200px|left|thumb|Figure 2: | + | [[Image:Triad.png |200px|left|thumb|Figure 2: HDAC8 utilizes a "catalytic dyad" of Asp and His. The His then assists in the coordination of the H<sub>2</sub>O nucleophile. This mechanism is reminiscent to the catalytic triad of Asp-His-Ser in many serine proteases (with H<sub>2</sub>O replacing the serine residue.]] |
===Binding Pocket=== | ===Binding Pocket=== | ||
| Line 28: | Line 28: | ||
At the rim of the active site, <scene name='81/811084/Asp101/5'>Asp101</scene> is involved in two hydrogen bonds between its own carbonyl oxygens and two consecutive amide hydrogens of incoming <scene name='81/811084/Ligand/7'>peptide-derived ligand</scene>. This forces the ligand to assume a cis-conformation. In addition, extensive interactions among many other polar atoms near the rim of the active site help keep the ligand lodged in the hydrophobic tunnel.<ref name="Vannini, A., Volpari, C., Gallinari, P.">Vannini, A., Volpari, C., Gallinari, P., Jones, P., Mattu, M., Carfí, A., ... & Di Marco, S. (2007). Substrate binding to histone deacetylases as shown by the crystal structure of the HDAC8–substrate complex. EMBO reports, 8(9), 879-884. https://doi.org/10.1038/sj.embor.7401047 </ref> | At the rim of the active site, <scene name='81/811084/Asp101/5'>Asp101</scene> is involved in two hydrogen bonds between its own carbonyl oxygens and two consecutive amide hydrogens of incoming <scene name='81/811084/Ligand/7'>peptide-derived ligand</scene>. This forces the ligand to assume a cis-conformation. In addition, extensive interactions among many other polar atoms near the rim of the active site help keep the ligand lodged in the hydrophobic tunnel.<ref name="Vannini, A., Volpari, C., Gallinari, P.">Vannini, A., Volpari, C., Gallinari, P., Jones, P., Mattu, M., Carfí, A., ... & Di Marco, S. (2007). Substrate binding to histone deacetylases as shown by the crystal structure of the HDAC8–substrate complex. EMBO reports, 8(9), 879-884. https://doi.org/10.1038/sj.embor.7401047 </ref> | ||
| - | ===Active site loop=== | + | ===Additional Features=== |
| + | |||
| + | ====Active site loop==== | ||
<scene name='81/811087/Active_site_loop_1_s30-k36/4'>N-Terminus L1 loop</scene>(amino acid residues 30-36) lines a large portion of one face of the active site pocket and extends to the protein surface. This results in a larger surface opening of the active site pocket. It is suggested that this loop has high flexibility, making the active site pocket opening highly malleable and able to change to accommodate binding to a variety of different ligands. <ref name="Somoza">Somoza J, Skene R. Structural snapshots of human HDAC8 provide insights into the class I histone deacetylases. Structure, 12(7), 1325-1334.2004. https://doi.org/10.1016/j.str.2004.04.012 </ref> | <scene name='81/811087/Active_site_loop_1_s30-k36/4'>N-Terminus L1 loop</scene>(amino acid residues 30-36) lines a large portion of one face of the active site pocket and extends to the protein surface. This results in a larger surface opening of the active site pocket. It is suggested that this loop has high flexibility, making the active site pocket opening highly malleable and able to change to accommodate binding to a variety of different ligands. <ref name="Somoza">Somoza J, Skene R. Structural snapshots of human HDAC8 provide insights into the class I histone deacetylases. Structure, 12(7), 1325-1334.2004. https://doi.org/10.1016/j.str.2004.04.012 </ref> | ||
| - | ===Ser 39=== | + | ====Ser 39==== |
The <scene name='81/811087/Active_site_loop_1_s30-k36/8'>Ser39</scene> has been shown to play a role in the activity of HDAC8 as the HDAC8 activity is regulated by phosphorylation at Ser39 by [https://en.wikipedia.org/wiki/Protein_kinase_A protein kinase A]. The phosphorylation of Ser39 leads to a decrease in the enzyme's activity. Ser39 lies at the surface of HDAC8, roughly 20 Å from the opening to the HDAC8 active site and it could be forming part of the surface that interacts with the target histone. The phosphorylation of Ser39 could disrupt the interaction between HDAC8 and the target histone. In addition, the phosphorylated Ser39 provokes a structural rearrangement near the active site by interacting with structural elements as Lys36, part of the conformational active loop L1, that extends into the active site. The Ser39 phosphorylation could therefore be inducing a conformation of the L1 loop that prohibits a competent substrate binding. <ref name="Somoza"> Somoza J, Skene R. Structural snapshots of human HDAC8 provide insights into the class I histone deacetylases. Structure, 12(7), 1325-1334.2004. https://doi.org/10.1016/j.str.2004.04.012 </ref> | The <scene name='81/811087/Active_site_loop_1_s30-k36/8'>Ser39</scene> has been shown to play a role in the activity of HDAC8 as the HDAC8 activity is regulated by phosphorylation at Ser39 by [https://en.wikipedia.org/wiki/Protein_kinase_A protein kinase A]. The phosphorylation of Ser39 leads to a decrease in the enzyme's activity. Ser39 lies at the surface of HDAC8, roughly 20 Å from the opening to the HDAC8 active site and it could be forming part of the surface that interacts with the target histone. The phosphorylation of Ser39 could disrupt the interaction between HDAC8 and the target histone. In addition, the phosphorylated Ser39 provokes a structural rearrangement near the active site by interacting with structural elements as Lys36, part of the conformational active loop L1, that extends into the active site. The Ser39 phosphorylation could therefore be inducing a conformation of the L1 loop that prohibits a competent substrate binding. <ref name="Somoza"> Somoza J, Skene R. Structural snapshots of human HDAC8 provide insights into the class I histone deacetylases. Structure, 12(7), 1325-1334.2004. https://doi.org/10.1016/j.str.2004.04.012 </ref> | ||
Revision as of 20:04, 9 April 2019
Histone Deacetylase 8 (HDAC 8)
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 1.4 Vannini, A., Volpari, C., Gallinari, P., Jones, P., Mattu, M., Carfí, A., ... & Di Marco, S. (2007). Substrate binding to histone deacetylases as shown by the crystal structure of the HDAC8–substrate complex. EMBO reports, 8(9), 879-884. https://doi.org/10.1038/sj.embor.7401047
- ↑ DesJarlais, R., & Tummino, P. J. (2016). Role of histone-modifying enzymes and their complexes in regulation of chromatin biology. Biochemistry, 55(11), 1584-1599. https://doi.org/10.1021/acs.biochem.5b01210
- ↑ 3.0 3.1 3.2 3.3 Somoza J, Skene R. Structural snapshots of human HDAC8 provide insights into the class I histone deacetylases. Structure, 12(7), 1325-1334.2004. https://doi.org/10.1016/j.str.2004.04.012
- ↑ Whitehead, L., Dobler, M. R., Radetich, B., Zhu, Y., Atadja, P. W., Claiborne, T., ... & Shao, W. (2011). Human HDAC isoform selectivity achieved via exploitation of the acetate release channel with structurally unique small molecule inhibitors. Bioorganic & medicinal chemistry, 19(15), 4626-4634. https://doi.org/10.1016/j.bmc.2011.06.030
- ↑ Seto, E., & Yoshida, M. (2014). Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harbor perspectives in biology, 6(4), a018713. https://doi.org/10.1101/cshperspect.a018713
- ↑ Eckschlager T, Plch, J, Stiborova M, Hrabeta J.Histone deacetylase inhibitors as anticancer drugs. International journal of molecular sciences, 18(7), 1414. 2017. https://dx.doi.org/10.3390%2Fijms18071414
- ↑ Vannini, A., Volpari, C., Filocamo, G., Casavola, E. C., Brunetti, M., Renzoni, D., ... & Steinkühler, C. (2004). Crystal structure of a eukaryotic zinc-dependent histone deacetylase, human HDAC8, complexed with a hydroxamic acid inhibitor. Proceedings of the National Academy of Sciences, 101(42), 15064-15069. https://dx.doi.org/10.1073%2Fpnas.0404603101