We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Emily Leiderman/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 23: | Line 23: | ||
=== Hydrophobic Pocket === | === Hydrophobic Pocket === | ||
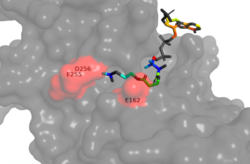
| - | [[Image:hydrophobic_pocket.png|250px|right|thumb|Figure 1. Image of the substrate Acetyl CoA bound in the groove of HAT1. The groove contains the hydrophobic pocket and is also contains the location of the active site.]] The active site consists of the Acetyl CoA ligand bound to the enzyme in a groove on the surface of the protein. The | + | [[Image:hydrophobic_pocket.png|250px|right|thumb|Figure 1. Image of the substrate Acetyl CoA bound in the groove of HAT1 (1BOB.pdb). The groove contains the hydrophobic pocket and is also contains the location of the active site.]] The active site consists of the Acetyl CoA ligand bound to the enzyme in a groove on the surface of the protein. The |
[https://en.wikipedia.org/wiki/Ligand ligand] is held in place by several bonds to protein residues that result in the formation of a <scene name='81/811098/Hydrophobic_pocket/1'>hydrophobic pocket</scene>. The hydrophobic pocket consists of the interacting side chains from residues | [https://en.wikipedia.org/wiki/Ligand ligand] is held in place by several bonds to protein residues that result in the formation of a <scene name='81/811098/Hydrophobic_pocket/1'>hydrophobic pocket</scene>. The hydrophobic pocket consists of the interacting side chains from residues | ||
<scene name='81/811098/Ile-217/3'>Ile-217</scene>, <scene name='81/811098/Pro-257/2'>Pro-257</scene>, | <scene name='81/811098/Ile-217/3'>Ile-217</scene>, <scene name='81/811098/Pro-257/2'>Pro-257</scene>, | ||
| Line 36: | Line 36: | ||
== Mechanism == | == Mechanism == | ||
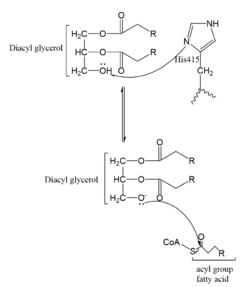
| - | Several mechanisms for the transfer of the acetyl group to the Lys-12 of the histone have been proposed. One possible mechanism consists of a 2-step process. The acetyl group is first transferred to a holding intermediate on the enzyme before subsequently being transferred to Lys-12. There are two cysteine residues, <scene name='81/811098/Cys-107/1'>Cys-107</scene> and <scene name='81/811098/Cys-234/1'>Cys-234</scene>, that could serve as sites to accept and acetyl group but are not in close enough proximity to the active site <ref name=Wu>PMID:22615379 </ref>. Another proposed mechanism that has been conserved across several HAT1 studies consists of deprotonating the | + | [[Image:Mechanism.PNG|250px|left|thumb|Figure 2. One possible acetylation mechanism of HAT1. Mechanism consists of deptronation of Lys-12 and then transfer of an acetyl group from Acetyl CoA to Lys-12.]]Several mechanisms for the transfer of the acetyl group to the Lys-12 of the histone have been proposed. One possible mechanism consists of a 2-step process. The acetyl group is first transferred to a holding intermediate on the enzyme before subsequently being transferred to Lys-12. There are two cysteine residues, <scene name='81/811098/Cys-107/1'>Cys-107</scene> and <scene name='81/811098/Cys-234/1'>Cys-234</scene>, that could serve as sites to accept and acetyl group but are not in close enough proximity to the active site <ref name=Wu>PMID:22615379 </ref>. Another proposed mechanism that has been conserved across several HAT1 studies consists of deprotonating the amino group on the Lys-12 of H4 (figure 2). This mechanism was proposed by Wu et al with respect to the human HAT1 protein. Analysis was done on the [https://www.rcsb.org/structure/1BOB 1BOB] model (retrieved from the protein database) and similar residues with respect to location and function were identified. Residues <scene name='81/811098/Glu-162/1'>Glu-162</scene>, <scene name='81/811098/Glu-255/1'>Glu-255</scene>, and <scene name='81/811098/Asp-256/1'>Asp-256</scene> cooperate to form a <scene name='81/811098/Catalytic_triad/2'>catalytic triad</scene> that establishes a basic environment that deprotonates the Lys-12 reside of H4 <ref name=Wu/>. Of those residues current research is uncertain which (if any) accept the proton of Lys-12. Deprotonation of the amino group on the Lys-12 makes the residue nucleophilic enough to directly attack the carbonyl carbon to initiate the acetyl transfer <ref name=Wu/>. The transfer mechanism is contingent on the conformational change and the formation of a functional gate that spans the concave groove over the bound Acetyl CoA and holds it in place while the catalytic [https://en.wikipedia.org/wiki/Deprotonation deprotonation] process takes place. |
| + | |||
| - | [[Image:Mechanism.PNG]] | ||
=== Protein Gate === | === Protein Gate === | ||
| Line 48: | Line 48: | ||
Homology of HAT1 to other proteins was shown by the analysis on similarity of secondary structural elements. The analysis showed that the Hat1 gene is conserved among other organisms, such as the chimpanzee, Rhesus monkey, dog, cow, mouse, rat, chicken, zebrafish, fruit fly, mosquito, [https://en.wikipedia.org/wiki/Caenorhabditis_elegans C. elegan], [https://en.wikipedia.org/wiki/Kluyveromyces_lactis K. lactis], [https://en.wikipedia.org/wiki/Eremothecium_gossypii E. gossypii], [https://en.wikipedia.org/wiki/Schizosaccharomyces_pombe S. pombe], [https://en.wikipedia.org/wiki/Magnaporthe_grisea M. oryzae], [https://en.wikipedia.org/wiki/Neurospora_crassa N. crassa], and [https://en.wikipedia.org/wiki/Arabidopsis_thaliana A. thaliana] <ref name=Dut/>. Overall, 269 organisms have orthologs to Hat1. These organisms showed similarity in aligned sequences over much of their length but had variability in the C-terminal domain. A possible reasoning for this is the idea that the C-terminal is not important for the HAT activity of Hat1 <ref name=Dut/>. Specifically, these organisms portrayed similar function in multiple histone targets, such as [https://en.wikipedia.org/wiki/Histone_H4 H4], H5, and H12 <ref name=Dut/>. | Homology of HAT1 to other proteins was shown by the analysis on similarity of secondary structural elements. The analysis showed that the Hat1 gene is conserved among other organisms, such as the chimpanzee, Rhesus monkey, dog, cow, mouse, rat, chicken, zebrafish, fruit fly, mosquito, [https://en.wikipedia.org/wiki/Caenorhabditis_elegans C. elegan], [https://en.wikipedia.org/wiki/Kluyveromyces_lactis K. lactis], [https://en.wikipedia.org/wiki/Eremothecium_gossypii E. gossypii], [https://en.wikipedia.org/wiki/Schizosaccharomyces_pombe S. pombe], [https://en.wikipedia.org/wiki/Magnaporthe_grisea M. oryzae], [https://en.wikipedia.org/wiki/Neurospora_crassa N. crassa], and [https://en.wikipedia.org/wiki/Arabidopsis_thaliana A. thaliana] <ref name=Dut/>. Overall, 269 organisms have orthologs to Hat1. These organisms showed similarity in aligned sequences over much of their length but had variability in the C-terminal domain. A possible reasoning for this is the idea that the C-terminal is not important for the HAT activity of Hat1 <ref name=Dut/>. Specifically, these organisms portrayed similar function in multiple histone targets, such as [https://en.wikipedia.org/wiki/Histone_H4 H4], H5, and H12 <ref name=Dut/>. | ||
| - | |||
| - | |||
| - | [[Image:Homology.PNG]] | ||
== References == | == References == | ||
Revision as of 22:41, 9 April 2019
Histone Acetyltransferase 1
| |||||||||||