User:Madeleine Wilson/Sandbox 1
From Proteopedia
| Line 7: | Line 7: | ||
===Histone Methylation=== | ===Histone Methylation=== | ||
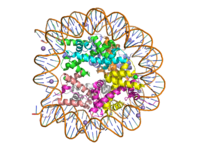
[[Image:human_nucleosome_ray_trace.png|200 px| right| thumb|Human nucleosome]] | [[Image:human_nucleosome_ray_trace.png|200 px| right| thumb|Human nucleosome]] | ||
| - | Histone proteins aid in the packing of DNA for the purpose of compacting the genome in the nucleus of the cell and regulating physical accessibility of genes for transcription. The protein itself is an octamer made of heterodimer core proteins H2a, H2b, H3, and H4, with H1 and H5 acting as linker proteins. About 145-157 base pairs wind around a histone core protein. | + | Histone proteins aid in the packing of DNA for the purpose of compacting the genome in the nucleus of the cell and regulating physical accessibility of genes for transcription. The protein itself is an octamer made of heterodimer core proteins H2a, H2b, H3, and H4, with H1 and H5 acting as linker proteins. About 145-157 base pairs wind around a histone core protein. <ref name="DesJarlais">PMID: 26745824</ref> Modifications to histone core proteins can affect the accessibility of genes in the genome and their ability to be transcribed. Some of these modifications include methylation/demethylation, acetylation/deacetylation, and ubiquitination/deubiquitination. (2) |
Specifically, histone methylation is associated with gene activation. (3) Many domain families fall under the Histone methylase family, one of these enzymes being the <scene name='81/811092/Set7_rotate/4'>SET7 domain</scene> family, which can target H3, H4, or H2a; each of these methylation sites can have different effects on gene expression within the genome. Typically, methylation of some of these sites are always present on both active and inactive genes, extra methylations required for activity. (4) Some tumor related genes such as p53 are site specifically methylated to promote biological function (5), whereas hypomethylation of CpG is linked to tumor genesis. (2) A particular enzyme in the SET7 domain family is lysine methyltransferase, which acts on the histone by adding a methyl group to Lys4 on H3; the addition results in promotion of gene unwinding and gene transcription. (4,3) | Specifically, histone methylation is associated with gene activation. (3) Many domain families fall under the Histone methylase family, one of these enzymes being the <scene name='81/811092/Set7_rotate/4'>SET7 domain</scene> family, which can target H3, H4, or H2a; each of these methylation sites can have different effects on gene expression within the genome. Typically, methylation of some of these sites are always present on both active and inactive genes, extra methylations required for activity. (4) Some tumor related genes such as p53 are site specifically methylated to promote biological function (5), whereas hypomethylation of CpG is linked to tumor genesis. (2) A particular enzyme in the SET7 domain family is lysine methyltransferase, which acts on the histone by adding a methyl group to Lys4 on H3; the addition results in promotion of gene unwinding and gene transcription. (4,3) | ||
| Line 37: | Line 37: | ||
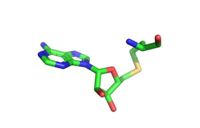
[[Image:Sinefugin.jpg|200px|left|thumb|Sinegungin]] | [[Image:Sinefugin.jpg|200px|left|thumb|Sinegungin]] | ||
Sinefungin is a potent methyltransferase inhibitor. It is a structural analog of S-adenosylmethionine that is more stable due to the ability to create two additional hydrogen bonds to its amine group in the active site. It has been used experimentally to inhibit the SET 7/9 protein on peritoneal fibrosis in mice and in human peritoneal mesothelial cells. Tamura et al. (2018) found that sinefungin suppressed the cell accumulation and thickening in methylglyoxal peritoneal fibrosis. | Sinefungin is a potent methyltransferase inhibitor. It is a structural analog of S-adenosylmethionine that is more stable due to the ability to create two additional hydrogen bonds to its amine group in the active site. It has been used experimentally to inhibit the SET 7/9 protein on peritoneal fibrosis in mice and in human peritoneal mesothelial cells. Tamura et al. (2018) found that sinefungin suppressed the cell accumulation and thickening in methylglyoxal peritoneal fibrosis. | ||
| + | |||
| + | </StructureSection> | ||
== References == | == References == | ||
| + | <references/> | ||
| + | |||
| - | 1. DesJarlais, R.; Tummino, P.J. Role of Histone-Modifying Enzymes and Their Complexes in Regulation of Chromatin Biology. Biochemistry 2016, 55: 1584-1599. | ||
2. Lu, D. Epigenetic modification enzymes: catalytic mechanisms and inhibitors. Acta Pharmaceutica Sinica B 2013, 3: 141-149. | 2. Lu, D. Epigenetic modification enzymes: catalytic mechanisms and inhibitors. Acta Pharmaceutica Sinica B 2013, 3: 141-149. | ||
Revision as of 01:32, 10 April 2019
Lysine Methyl Transferase, Homo Sapiens
| |||||||||||
References
- ↑ DesJarlais R, Tummino PJ. Role of Histone-Modifying Enzymes and Their Complexes in Regulation of Chromatin Biology. Biochemistry. 2016 Mar 22;55(11):1584-99. doi: 10.1021/acs.biochem.5b01210. Epub , 2016 Jan 26. PMID:26745824 doi:http://dx.doi.org/10.1021/acs.biochem.5b01210
2. Lu, D. Epigenetic modification enzymes: catalytic mechanisms and inhibitors. Acta Pharmaceutica Sinica B 2013, 3: 141-149.
3. Dong, X.; Weng, Z. The correlation between histone modifications and gene expression. Epigenomics 2013, 5: 113-6.
4. Xiao, B.; Jing, C.; Wilson, J.; Walker, P.; Vasisht, N.; et al. Structure and catalytic mechanism of the human histone methyltranferase SET7/9. Letters to Nature 2003, 412: 652-655.
5. Del Rizzo, P. A.; Trievel, R.C. Substrate and product specificities of SET domain methyltransferases. Epigenetics 2011, 6: 1059-1067
6. Mao, X.; Qiao, Z.; Fan, C.; Guo, A.; Yu, X.; Jin, F. Expression pattern and methylation of estrogen receptor α in breast intraductal proliferative lesions. Oncology Reports 2016, 36: 1868-1874.
7. Tamura, R., Doi, S., Nakashima, A., Sasaki, K., Maeda, K., Ueno, T., & Masaki, T. (2018). Inhibition of the H3K4 methyltransferase SET7/9 ameliorates peritoneal fibrosis. PloS one, 13(5), e0196844. doi:10.1371/journal.pone.0196844
8. Schluckebier et al. (1997), Differential binding of S-andeosylmethionine S-adenosylhomocysteine and Sinefungin to adenine-specific DNA methyltransferase M. TaqI; J. Mol. Biol., 265 56