We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Madeleine Wilson/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
=Lysine Methyl Transferase, ''Homo Sapiens''= | =Lysine Methyl Transferase, ''Homo Sapiens''= | ||
| + | |||
<StructureSection load='1O9S' size='350' frame='true' side='right' caption='Lysine Methyl Transferase' scene=’C_terminal_domain’> | <StructureSection load='1O9S' size='350' frame='true' side='right' caption='Lysine Methyl Transferase' scene=’C_terminal_domain’> | ||
| - | <references/> | ||
== Introduction == | == Introduction == | ||
| Line 9: | Line 9: | ||
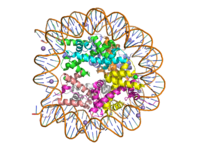
Histone proteins aid in the packing of DNA for the purpose of compacting the genome in the nucleus of the cell and regulating physical accessibility of genes for transcription. The protein itself is an octamer made of heterodimer core proteins H2a, H2b, H3, and H4, with H1 and H5 acting as linker proteins. About 145-157 base pairs wind around a histone core protein. <ref name="DesJarlais">PMID: 26745824</ref> Modifications to histone core proteins can affect the accessibility of genes in the genome and their ability to be transcribed. Some of these modifications include methylation/demethylation, acetylation/deacetylation, and ubiquitination/deubiquitination. <ref name="Lun">DOI: 10.1016/j.apsb.2013.04.007</ref> | Histone proteins aid in the packing of DNA for the purpose of compacting the genome in the nucleus of the cell and regulating physical accessibility of genes for transcription. The protein itself is an octamer made of heterodimer core proteins H2a, H2b, H3, and H4, with H1 and H5 acting as linker proteins. About 145-157 base pairs wind around a histone core protein. <ref name="DesJarlais">PMID: 26745824</ref> Modifications to histone core proteins can affect the accessibility of genes in the genome and their ability to be transcribed. Some of these modifications include methylation/demethylation, acetylation/deacetylation, and ubiquitination/deubiquitination. <ref name="Lun">DOI: 10.1016/j.apsb.2013.04.007</ref> | ||
| - | Specifically, histone methylation is associated with gene activation. <ref name="Dong">PMID: 23566087</ref> Many domain families fall under the Histone methylase family, one of these enzymes being the <scene name='81/811092/Set7_rotate/4'>SET7 domain</scene> family, which can target H3, H4, or H2a; each of these methylation sites can have different effects on gene expression within the genome. Typically, methylation of some of these sites are always present on both active and inactive genes, extra methylations required for activity. <ref name="Xiao">doi:10.1038/nature01378</ref> Some tumor related genes such as p53 are site specifically methylated to promote biological function <ref name="Rizzo">PMID: 21847010</ref>, whereas hypomethylation of CpG is linked to tumor genesis. <ref name="Lun" /> A particular enzyme in the SET7 domain family is lysine methyltransferase, which acts on the histone by adding a methyl group to Lys4 on H3; the addition results in promotion of gene unwinding and gene transcription. <ref name="Xiao" />,<ref name="Dong"> | + | Specifically, histone methylation is associated with gene activation. <ref name="Dong">PMID: 23566087</ref> Many domain families fall under the Histone methylase family, one of these enzymes being the <scene name='81/811092/Set7_rotate/4'>SET7 domain</scene> family, which can target H3, H4, or H2a; each of these methylation sites can have different effects on gene expression within the genome. Typically, methylation of some of these sites are always present on both active and inactive genes, extra methylations required for activity. <ref name="Xiao">doi:10.1038/nature01378</ref> Some tumor related genes such as p53 are site specifically methylated to promote biological function <ref name="Rizzo">PMID: 21847010</ref>, whereas hypomethylation of CpG is linked to tumor genesis. <ref name="Lun" /> A particular enzyme in the SET7 domain family is lysine methyltransferase, which acts on the histone by adding a methyl group to Lys4 on H3; the addition results in promotion of gene unwinding and gene transcription. <ref name="Xiao" />, <ref name="Dong" /> |
==KMT Structure== | ==KMT Structure== | ||
| Line 22: | Line 22: | ||
===The C-Terminal Domain=== | ===The C-Terminal Domain=== | ||
| - | |||
The <scene name='81/811091/C_terminal_domain/1'>C-terminal segment</scene> of lysine methyltransferase is essential for the catalytic activity of the enzyme. Hydrophobic packing of the C-terminal segment (residues 345-366) forms the lysine access channel. Residues 337-349 create a <scene name='81/811086/Beta_hairpin/2'>beta hairpin structure</scene> that stabilizes the orientation of two tyrosine residues Tyr 335 and Tyr337 that form the lysine access channel. The hydrophobic packing of the C-terminal <scene name='81/811091/C_terminal_domain/7'>alpha helix</scene> against beta sheet 19 (specifically residue 299) orient the SAM cofactor so the methyl donating group is oriented toward the lysine access channel. donating group is oriented toward the lysine access channel. | The <scene name='81/811091/C_terminal_domain/1'>C-terminal segment</scene> of lysine methyltransferase is essential for the catalytic activity of the enzyme. Hydrophobic packing of the C-terminal segment (residues 345-366) forms the lysine access channel. Residues 337-349 create a <scene name='81/811086/Beta_hairpin/2'>beta hairpin structure</scene> that stabilizes the orientation of two tyrosine residues Tyr 335 and Tyr337 that form the lysine access channel. The hydrophobic packing of the C-terminal <scene name='81/811091/C_terminal_domain/7'>alpha helix</scene> against beta sheet 19 (specifically residue 299) orient the SAM cofactor so the methyl donating group is oriented toward the lysine access channel. donating group is oriented toward the lysine access channel. | ||
===The Active Site=== | ===The Active Site=== | ||
| - | |||
The active site and binding pocket of KMT have several essential characteristics for the overall efficiency. First, the lysine of the histone enters the active site “with difficulty” which is facilitated by the faces of flanking <scene name='81/811092/Tyrosine_channel_2/1'>tyrosines (Y35, Y37)</scene>. Once in the active site, the alkyl part of the histone chain is stabilized by the <scene name='81/811092/Hydrophobic_binding_pocket/1'>hydrophobic binding pocket</scene>, and polar residues are stabilized by hydrogen bonding interactions on the surface. The Y335 and Y337 are also essential for stabilization of histone chain via hydrogen bonding. | The active site and binding pocket of KMT have several essential characteristics for the overall efficiency. First, the lysine of the histone enters the active site “with difficulty” which is facilitated by the faces of flanking <scene name='81/811092/Tyrosine_channel_2/1'>tyrosines (Y35, Y37)</scene>. Once in the active site, the alkyl part of the histone chain is stabilized by the <scene name='81/811092/Hydrophobic_binding_pocket/1'>hydrophobic binding pocket</scene>, and polar residues are stabilized by hydrogen bonding interactions on the surface. The Y335 and Y337 are also essential for stabilization of histone chain via hydrogen bonding. | ||
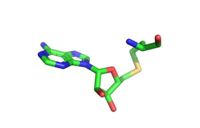
The <scene name='81/811092/Active_site_w_water/2'>active site</scene> itself contains the cofactor S-adenosyl methionine (SAM) which donates the methyl group in the reaction. <ref name="Xiao" /> | The <scene name='81/811092/Active_site_w_water/2'>active site</scene> itself contains the cofactor S-adenosyl methionine (SAM) which donates the methyl group in the reaction. <ref name="Xiao" /> | ||
[[Image:AdoHcy.jpg|200px|right|thumb|S-adenosyl homocysteine (SAH)]] | [[Image:AdoHcy.jpg|200px|right|thumb|S-adenosyl homocysteine (SAH)]] | ||
The reaction is catalyzed by Y305, Y245, carbonyl oxygens of the main chain in residues 295 and 290. Y305 and the carbonyl oxygens stabilize and pull electron density off a water to pull on one of the hydrogens off the nitrogen of the lysine, while oxygen of Y245 pulls on the other hydrogen of the nitrogen. Both of these actions allow nitrogen to become more nucleophilic and attack the carbon of the methyl group on the SAM, which is attached to a positively charged sulfur. The methyl group is then transferred and the sulfur is neutral; SAM has been converted to S-adenosyl homocysteine (SAH). <ref name="Xiao" /> | The reaction is catalyzed by Y305, Y245, carbonyl oxygens of the main chain in residues 295 and 290. Y305 and the carbonyl oxygens stabilize and pull electron density off a water to pull on one of the hydrogens off the nitrogen of the lysine, while oxygen of Y245 pulls on the other hydrogen of the nitrogen. Both of these actions allow nitrogen to become more nucleophilic and attack the carbon of the methyl group on the SAM, which is attached to a positively charged sulfur. The methyl group is then transferred and the sulfur is neutral; SAM has been converted to S-adenosyl homocysteine (SAH). <ref name="Xiao" /> | ||
| - | |||
==Inhibitors== | ==Inhibitors== | ||
| Line 42: | Line 39: | ||
<references/> | <references/> | ||
| - | + | ==Student Contributors== | |
| - | + | Lauryn Padgett, | |
| - | + | Alexandra Pentala, | |
| - | + | Madeleine Wilson | |
Revision as of 01:53, 10 April 2019
Lysine Methyl Transferase, Homo Sapiens
| |||||||||||
References
- ↑ DesJarlais R, Tummino PJ. Role of Histone-Modifying Enzymes and Their Complexes in Regulation of Chromatin Biology. Biochemistry. 2016 Mar 22;55(11):1584-99. doi: 10.1021/acs.biochem.5b01210. Epub , 2016 Jan 26. PMID:26745824 doi:http://dx.doi.org/10.1021/acs.biochem.5b01210
- ↑ 2.0 2.1 doi: https://dx.doi.org/10.1016/j.apsb.2013.04.007
- ↑ 3.0 3.1 Dong X, Weng Z. The correlation between histone modifications and gene expression. Epigenomics. 2013 Apr;5(2):113-6. doi: 10.2217/epi.13.13. PMID:23566087 doi:http://dx.doi.org/10.2217/epi.13.13
- ↑ 4.0 4.1 4.2 4.3 Xiao B, Jing C, Wilson JR, Walker PA, Vasisht N, Kelly G, Howell S, Taylor IA, Blackburn GM, Gamblin SJ. Structure and catalytic mechanism of the human histone methyltransferase SET7/9. Nature. 2003 Feb 6;421(6923):652-6. Epub 2003 Jan 22. PMID:12540855 doi:10.1038/nature01378
- ↑ Del Rizzo PA, Trievel RC. Substrate and product specificities of SET domain methyltransferases. Epigenetics. 2011 Sep 1;6(9):1059-67. doi: 10.4161/epi.6.9.16069. Epub 2011 Sep, 1. PMID:21847010 doi:http://dx.doi.org/10.4161/epi.6.9.16069
- ↑ Schluckebier G, Kozak M, Bleimling N, Weinhold E, Saenger W. Differential binding of S-adenosylmethionine S-adenosylhomocysteine and Sinefungin to the adenine-specific DNA methyltransferase M.TaqI. J Mol Biol. 1997 Jan 10;265(1):56-67. PMID:8995524 doi:http://dx.doi.org/10.1006/jmbi.1996.0711
- ↑ Tamura R, Doi S, Nakashima A, Sasaki K, Maeda K, Ueno T, Masaki T. Inhibition of the H3K4 methyltransferase SET7/9 ameliorates peritoneal fibrosis. PLoS One. 2018 May 3;13(5):e0196844. doi: 10.1371/journal.pone.0196844., eCollection 2018. PMID:29723250 doi:http://dx.doi.org/10.1371/journal.pone.0196844
Student Contributors
Lauryn Padgett, Alexandra Pentala, Madeleine Wilson