User:Caitlin Marie Gaich/Sandbox1
From Proteopedia
(Difference between revisions)
| Line 2: | Line 2: | ||
<StructureSection load='4psw' size='350' frame='true' side='right' caption='HAT1/HAT2 Complex pdb: 4PSW'> | <StructureSection load='4psw' size='350' frame='true' side='right' caption='HAT1/HAT2 Complex pdb: 4PSW'> | ||
== Introduction == | == Introduction == | ||
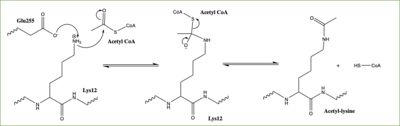
| - | [https://en.wikipedia.org/wiki/Histone Histones] | + | [https://en.wikipedia.org/wiki/Histone Histones]are essential for proper DNA packaging and are the key building blocks of [https://en.wikipedia.org/wiki/Chromatin chromatin]. They are subject to post-translational modifications and play important roles in replication, transcription, heterochromatin maintenance, and DNA repair. Histones can be modified in a variety of ways, including: methylations, demethylation, acetylation, and deacetylation, all leading to either the condensation or relaxation of DNA and as a consequence turning on or off DNA transcription. Histone acetylation is a common histone modification. This involves the transfer of an acetyl moiety from Acetyl Coenzyme A (AcCoA) to an ε-amino group of the target lysine residue on a histone. This reaction is catalyzed by the histone acetyltransferase (HAT) enzyme families. The specific histone acetylation modification is an important [https://en.wikipedia.org/wiki/Epigenetics epigenetic] marker. It plays a role in RNA synthesis and there a known correlation between gene activity and histone acetylation. Any misregulations of the HAT enzyme can possibly lead to cancer, cardiovascular disease, and HIV. |
== HAT1 Background == | == HAT1 Background == | ||
| Line 17: | Line 17: | ||
The acetyl-CoA HAT1 active site is parallel to the C-terminal domain of the HAT1 protein. Acetyl-CoA fits structurally into the small binding site due to the kinked pantetheine group giving the molecule a bent confirmation. Once bound, most of the acetyl-CoA molecule is <scene name='81/811717/Tight_protein-ligand_intxn/1'>buried in the protein</scene> (~60%). Hydrophobic contacts, hydrogen bonds, and salt bridges help to stabilize the protein-ligand interaction. HAT1 protein-ligand contact is concentrated in three areas: C-terminal end of helix alpha 7, C terminal end of strand beta-14/loop Beta15-Alpha9, and N-terminal half of helix alpha 9 <ref name=”Dutnall”>PMID:10384314</ref>. | The acetyl-CoA HAT1 active site is parallel to the C-terminal domain of the HAT1 protein. Acetyl-CoA fits structurally into the small binding site due to the kinked pantetheine group giving the molecule a bent confirmation. Once bound, most of the acetyl-CoA molecule is <scene name='81/811717/Tight_protein-ligand_intxn/1'>buried in the protein</scene> (~60%). Hydrophobic contacts, hydrogen bonds, and salt bridges help to stabilize the protein-ligand interaction. HAT1 protein-ligand contact is concentrated in three areas: C-terminal end of helix alpha 7, C terminal end of strand beta-14/loop Beta15-Alpha9, and N-terminal half of helix alpha 9 <ref name=”Dutnall”>PMID:10384314</ref>. | ||
| - | The β-methyl of the acetyl group interactions in the hydrophobic pocket formed by the side chain of residues: <scene name='81/811717/Hydrophobic_pocket/2'>Ile-217,Pro-257, Phe-261</scene>. The <scene name='81/811717/Phe_interaction/ | + | The β-methyl of the acetyl group interactions in the hydrophobic pocket formed by the side chain of residues: <scene name='81/811717/Hydrophobic_pocket/2'>Ile-217,Pro-257, Phe-261</scene>. The <scene name='81/811717/Phe_interaction/2'>carbonyl oxygen of the acetyl group hydrogen bonds with the aminde of the main chain Phe-220</scene> and the <scene name='81/811717/Asn_ligand_interaction/1'>sulfur of the acetyl-group interacts, as a hydrogen bond, with Asn-258</scene>. These interactions keep acetyl-CoA in the correct position of the active site for the transfer of the acetyl-group. |
Revision as of 21:17, 12 April 2019
Histone Acetyltransferase HAT1/HAT2 Complex, Saccharomyces cerevisiae
| |||||||||||