Introduction
Histone Methylation
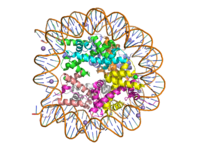
Human nucleosome particle, pbd code: 5y0c
Histone proteins aid in the packing of DNA for the purpose of compacting the genome in the nucleus of the cell and regulating physical accessibility of genes for transcription. The protein itself is an octamer of core proteins H2a, H2b, H3, and H4, which organize into two heterodimers; H1 and H5 act as linker proteins. About 145-157 base pairs wind around a histone heterodimer core. [1] Modifications to histone core proteins can affect the accessibility of transcription factors in the genome, either promoting or inhibiting transcription. Some of these modifications include methylation/demethylation, acetylation/deacetylation, and ubiquitination/deubiquitination. [2]
Specifically, histone methylation is associated with gene activation. [3] Many domain families fall under the histone methylase family, one of these enzymes being the family, which can target H3, H4, or H2a. Sites known for gene activation are Lysine-4, Lys-36, and Lys-79 on H3; whereas, methylation at Lys-9 and Lys- 27 on H3 and Lys-20 on H4 are known for gene inactivation.[4] Typically, methylation of some of these sites are always present on both active and inactive genes, extra methylations required for activity; specifics of this characteristic depend on site and species of organism. [5] Some tumor related genes such as p53 are site specifically methylated to promote biological function [4], whereas hypomethylation of CpG is linked to tumor genesis. [2] A particular enzyme in the SET7 domain family is lysine methyltransferase, which acts on the histone by adding a methyl group to Lys4 on H3; the addition results in promotion of gene unwinding and gene transcription. [5], [3]
Lysine Methyltransferase (KMT) Structure
Composition
The secondary structure is composed of 10% and 37% . The helical composition includes 3 , with two residing in the SET domain and one in the C-terminal domain. The alpha helices in the SET domain are two turns while the C-terminal helix is by far the largest with 4 turns. There are also 2 in the SET domain which are each one turn. There are 21 total beta strands which reside in the N-terminal domain and the SET domain. The beta strands are primarily anti-parallel and multiple antiparallel strands are connected by and beta turns.
The N-terminal Domain

Clustal alignment of N-terminals
The N-terminal domain has no notable function in regard to the activity of KMT; however, it has been related to stability of the enzyme and most likely prevents its degradation within the cell. The N-terminal domain is highly conserved across species, which supports of the claim of necessity for stability. A Clustal alignment of a portion of the region shows a variety of high level species with conservation.
The C-Terminal Domain
The of lysine methyltransferase is essential for the catalytic activity of the enzyme. Hydrophobic packing of the C-terminal segment (residues 345-366) forms the lysine access channel. Residues 337-349 create a that stabilizes the orientation of two tyrosine residues Tyr 335 and Tyr337 that form the lysine access channel. The hydrophobic packing of the C-terminal against beta sheet 19 (specifically residue 299) orient the SAM cofactor so the methyl donating group is oriented toward the lysine access channel. donating group is oriented toward the lysine access channel.
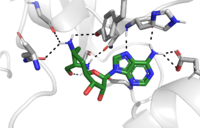
The Active Site
The active site and binding pocket of KMT have several essential characteristics for the overall efficiency. First, the lysine of the histone enters the active site “with difficulty” which is facilitated by the faces of flanking , also known as the lysine access channel. Once in the active site, the alkyl part of the histone chain is stabilized by the , and polar residues are stabilized by hydrogen bonding interactions on the surface. The Y335 and Y337 are also essential for stabilization of histone chain via hydrogen bonding.
The itself contains the cofactor S-adenosyl methionine (SAM) which donates the methyl group in the reaction. [5]
The reaction is catalyzed by Y305, Y245, carbonyl oxygens of the main chain in residues 295 and 290. Y305 and the carbonyl oxygens stabilize and pull electron density off a water to pull on one of the hydrogens off the nitrogen of the lysine, while oxygen of Y245 pulls on the other hydrogen of the nitrogen. Both of these actions allow nitrogen to become more nucleophilic and attack the carbon of the methyl group on the SAM, which is attached to a positively charged sulfur. The methyl group is then transferred and the sulfur is neutral; SAM has been converted to S-adenosyl homocysteine (SAH). [5]
Inhibitors
Sinefungin is a potent methyltransferase inhibitor. It is a structural analog of S-adenosylmethionine that is more stable due to the ability to create two additional hydrogen bonds to its amine group in the active site. It has been used experimentally to inhibit the SET 7/9 protein on peritoneal fibrosis in mice and in human peritoneal mesothelial cells. [6] Tamura et al. (2018) found that sinefungin suppressed the cell accumulation and thickening in methylglyoxal peritoneal fibrosis. [7]