We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Sean Callahan/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 5: | Line 5: | ||
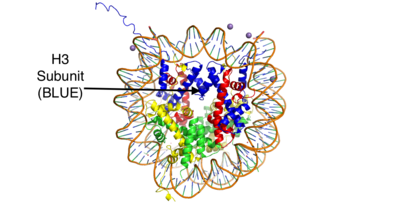
Histones are positively charged proteins that help organize DNA into tightly packed chromosomes by acting as a spool for DNA to wrap around. Histones are composed of 4 subunits (H2A, H2B, H3, and H4) and have the capability to either loosen their interaction with DNA to promote transcription or tighten their interaction to suppress transcription. Some of the mechanisms that histones use to achieve these different interactions include the addition or removal of acetyl, methyl, or phosphate groups. Demethylases are responsible for removing methyl groups from different histone residues. While this is typically associated with increasing histone-DNA interaction, and thus silencing transcription, demethylation has also been associated with the promotion of transcription depending on the residue that is being demethylated.[[Image:HistoneStructure.png | 400px| right| thumb| Figure 1: This is the crystal structure of a histone bound to DNA. Its subunits are color coded.]] | Histones are positively charged proteins that help organize DNA into tightly packed chromosomes by acting as a spool for DNA to wrap around. Histones are composed of 4 subunits (H2A, H2B, H3, and H4) and have the capability to either loosen their interaction with DNA to promote transcription or tighten their interaction to suppress transcription. Some of the mechanisms that histones use to achieve these different interactions include the addition or removal of acetyl, methyl, or phosphate groups. Demethylases are responsible for removing methyl groups from different histone residues. While this is typically associated with increasing histone-DNA interaction, and thus silencing transcription, demethylation has also been associated with the promotion of transcription depending on the residue that is being demethylated.[[Image:HistoneStructure.png | 400px| right| thumb| Figure 1: This is the crystal structure of a histone bound to DNA. Its subunits are color coded.]] | ||
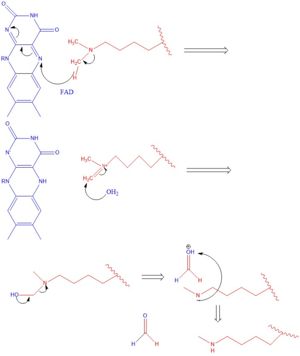
| - | There are two main classes of demethylases, and they are categorized by their co-factors and co-substrates. One class of demethylases uses an [https://en.wikipedia.org/wiki/Flavin_adenine_dinucleotide | + | There are two main classes of demethylases, and they are categorized by their co-factors and co-substrates. One class of demethylases uses an [https://en.wikipedia.org/wiki/Flavin_adenine_dinucleotide FAD] co-factor to catalyze the demethylation reaction. The other class of demethylases uses a Fe2+ ion and [https://en.wikipedia.org/wiki/Alpha-Ketoglutaric_acid α-ketoglutarate] as a co-substrate to catalyze the reaction. Although the co-factors used are different, both classes operate by hydroxylating the target methyl group. Lysine Specific Demethylases 1 is a histone demethylase that uses FAD as a co-factor<ref name="Forneris">PMID: 15811342</ref>. Specifically, LSD1 is responsible for demethylating Lys 4 and Lys 9 on the H3 subunit of the histone. |
==Structure== | ==Structure== | ||
Revision as of 17:29, 19 April 2019
Lysine Specific Demethylase 1 (Homo sapiens)
| |||||||||||
References
- ↑ 1.0 1.1 Forneris F, Binda C, Vanoni MA, Mattevi A, Battaglioli E. Histone demethylation catalysed by LSD1 is a flavin-dependent oxidative process. FEBS Lett. 2005 Apr 11;579(10):2203-7. doi: 10.1016/j.febslet.2005.03.015. PMID:15811342 doi:http://dx.doi.org/10.1016/j.febslet.2005.03.015
- ↑ 2.0 2.1 Da G, Lenkart J, Zhao K, Shiekhattar R, Cairns BR, Marmorstein R. Structure and function of the SWIRM domain, a conserved protein module found in chromatin regulatory complexes. Proc Natl Acad Sci U S A. 2006 Feb 14;103(7):2057-62. Epub 2006 Feb 3. PMID:16461455
- ↑ 3.0 3.1 3.2 3.3 3.4 Stavropoulos P, Blobel G, Hoelz A. Crystal structure and mechanism of human lysine-specific demethylase-1. Nat Struct Mol Biol. 2006 Jul;13(7):626-32. Epub 2006 Jun 25. PMID:16799558 doi:10.1038/nsmb1113
- ↑ 4.0 4.1 Abdel-Magid AF. Lysine-Specific Demethylase 1 (LSD1) Inhibitors as Potential Treatment for Different Types of Cancers. ACS Med Chem Lett. 2017 Oct 27;8(11):1134-1135. doi:, 10.1021/acsmedchemlett.7b00426. eCollection 2017 Nov 9. PMID:29152043 doi:http://dx.doi.org/10.1021/acsmedchemlett.7b00426
- ↑ Forneris F, Binda C, Vanoni MA, Battaglioli E, Mattevi A. Human histone demethylase LSD1 reads the histone code. J Biol Chem. 2005 Dec 16;280(50):41360-5. doi: 10.1074/jbc.M509549200. Epub 2005 , Oct 13. PMID:16223729 doi:http://dx.doi.org/10.1074/jbc.M509549200
- ↑ Qian C, Zhang Q, Li S, Zeng L, Walsh MJ, Zhou MM. Structure and chromosomal DNA binding of the SWIRM domain. Nat Struct Mol Biol. 2005 Dec;12(12):1078-85. Epub 2005 Nov 20. PMID:16299514 doi:10.1038/nsmb1022