User:Asif Hossain/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 12: | Line 12: | ||
==HDAC8 Structure== | ==HDAC8 Structure== | ||
| - | The HDAC8 comprises a single α/β domain that is composed of an one <scene name='81/811084/Beta_sheets/ | + | The HDAC8 comprises a single α/β domain that is composed of an one <scene name='81/811084/Beta_sheets/4'>β-sheet</scene> with eight parallel β-strands sandwiched between 13 <scene name='81/811084/Alpha_helicesv2/3'>α-helices</scene>. The HDAC8 consists of 377 amino acids, half of which are contained in the secondary structure elements and the other half are contained in loops that link the various elements of the secondary structure. The residues forming active site and catalytic machinery of the enzyme is found in the loops from the C-terminal ends of the strands of the core β-sheet. <ref name="Somoza"> Somoza J, Skene R. Structural snapshots of human HDAC8 provide insights into the class I histone deacetylases. Structure, 12(7), 1325-1334.2004. https://doi.org/10.1016/j.str.2004.04.012 </ref> |
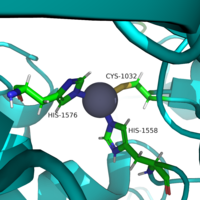
===Zinc Ion=== | ===Zinc Ion=== | ||
| Line 31: | Line 31: | ||
<scene name='81/811087/Active_site_loop_1_s30-k36/11'>N-Terminus L1 loop</scene>(amino acid residues 30-36) lines a large part of one face of the active site pocket and extends to the protein surface. This results in a larger surface opening of the active site pocket. It is suggested that this loop has high flexibility that enables HDAC8 to more efficiently accommodate binding to a variety of different ligands. <ref name="Somoza">Somoza J, Skene R. Structural snapshots of human HDAC8 provide insights into the class I histone deacetylases. Structure, 12(7), 1325-1334.2004. https://doi.org/10.1016/j.str.2004.04.012 </ref> | <scene name='81/811087/Active_site_loop_1_s30-k36/11'>N-Terminus L1 loop</scene>(amino acid residues 30-36) lines a large part of one face of the active site pocket and extends to the protein surface. This results in a larger surface opening of the active site pocket. It is suggested that this loop has high flexibility that enables HDAC8 to more efficiently accommodate binding to a variety of different ligands. <ref name="Somoza">Somoza J, Skene R. Structural snapshots of human HDAC8 provide insights into the class I histone deacetylases. Structure, 12(7), 1325-1334.2004. https://doi.org/10.1016/j.str.2004.04.012 </ref> | ||
| - | The <scene name='81/811087/Active_site_loop_1_s30-k36/ | + | The activity of HDAC8 is regulated by phosphorylation at <scene name='81/811087/Active_site_loop_1_s30-k36/12'>Ser39</scene> by [https://en.wikipedia.org/wiki/Protein_kinase_A protein kinase A(PKA)] as the phosphorylation has been shown to decrease the enzyme’s activity.<ref name="Somoza">Somoza J, Skene R. Structural snapshots of human HDAC8 provide insights into the class I histone deacetylases. Structure, 12(7), 1325-1334.2004. https://doi.org/10.1016/j.str.2004.04.012 </ref> Ser39 lies at the surface of HDAC8, approximately 20Å from the opening to the HDAC8 active site so it is likely part of the surface that interacts with the target histone. The interaction between the HDAC8 and target histone could be disrupted by the phosphorylation of Ser39 because the phosphorylated Ser39 elicits a structural rearrangement that extends to the active site and thus disrupts enzyme activity. Ser39 elicits this rearrangement by interactions with structural elements in the conformational active loop L1, such as K36. Therefore, the Ser39 phosphorylation by PKA is likely inducing a conformational change of the L1 loop that prohibits a competent substrate binding.<ref name="Somoza">Somoza J, Skene R. Structural snapshots of human HDAC8 provide insights into the class I histone deacetylases. Structure, 12(7), 1325-1334.2004. https://doi.org/10.1016/j.str.2004.04.012 </ref> |
| + | |||
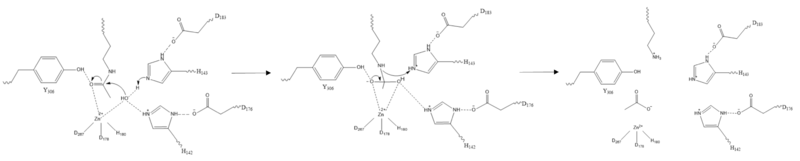
==Mechanism== | ==Mechanism== | ||
Revision as of 19:41, 23 April 2019
Histone Deacetylase 8 (HDAC 8)
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 Vannini, A., Volpari, C., Gallinari, P., Jones, P., Mattu, M., Carfí, A., ... & Di Marco, S. (2007). Substrate binding to histone deacetylases as shown by the crystal structure of the HDAC8–substrate complex. EMBO reports, 8(9), 879-884. https://doi.org/10.1038/sj.embor.7401047
- ↑ DesJarlais, R., & Tummino, P. J. (2016). Role of histone-modifying enzymes and their complexes in regulation of chromatin biology. Biochemistry, 55(11), 1584-1599. https://doi.org/10.1021/acs.biochem.5b01210
- ↑ 3.0 3.1 3.2 3.3 3.4 Somoza J, Skene R. Structural snapshots of human HDAC8 provide insights into the class I histone deacetylases. Structure, 12(7), 1325-1334.2004. https://doi.org/10.1016/j.str.2004.04.012
- ↑ Whitehead, L., Dobler, M. R., Radetich, B., Zhu, Y., Atadja, P. W., Claiborne, T., ... & Shao, W. (2011). Human HDAC isoform selectivity achieved via exploitation of the acetate release channel with structurally unique small molecule inhibitors. Bioorganic & medicinal chemistry, 19(15), 4626-4634. https://doi.org/10.1016/j.bmc.2011.06.030
- ↑ Seto, E., & Yoshida, M. (2014). Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harbor perspectives in biology, 6(4), a018713. https://doi.org/10.1101/cshperspect.a018713
- ↑ Eckschlager T, Plch, J, Stiborova M, Hrabeta J.Histone deacetylase inhibitors as anticancer drugs. International journal of molecular sciences, 18(7), 1414. 2017. https://dx.doi.org/10.3390%2Fijms18071414
- ↑ Vannini, A., Volpari, C., Filocamo, G., Casavola, E. C., Brunetti, M., Renzoni, D., ... & Steinkühler, C. (2004). Crystal structure of a eukaryotic zinc-dependent histone deacetylase, human HDAC8, complexed with a hydroxamic acid inhibitor. Proceedings of the National Academy of Sciences, 101(42), 15064-15069. https://dx.doi.org/10.1073%2Fpnas.0404603101