Histones
Histones are proteins found in the nucleus that are the key building blocks of chromatin and are essential for proper DNA packaging and transcription. In the first step of DNA packaging, two copies of the four core histone proteins (H1A, H2A, H3, and H4) form an octamer in which DNA directly interacts with and wraps around, forming the nucleosome. 20-24% of residues making up the histone octamer are arginine and lysine, causing a net positive charge, especially at the outer surfaces of the histone core where negatively-charged DNA is bound [1] (Figure 1). After translation, the positively charged tails of the histone core that are often subject to modifications, such as acetylation or methylation. These modifications can regulate the processes of DNA repair, replication, transcription, and heterochromatin maintenance.
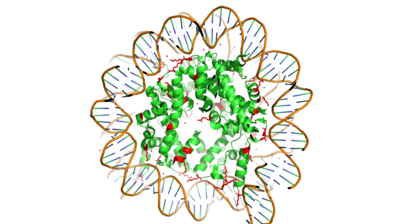
Figure 1. Nucleosome consisting of the Histone core & DNA bound
Histone Modification
Histones can be modified in a variety of ways, including: methylation, demethylation, acetylation, deacetylation and many others, all leading to either the condensation or relaxation of DNA and as a consequence turning on or off DNA transcription. Histone acetylation is a common histone modification that involves the transfer of an acetyl moiety from Acetyl Coenzyme A (AcCoA) to an ε-amino group of the target lysine residue on a histone. This reaction is catalyzed by various histone acetyltransferase (HAT) enzymes. The specific histone acetylation modification is an important epigenetic marker. It plays a role in RNA synthesis and there a known correlation between gene activity and histone acetylation. Any misregulations of the HAT enzyme can possibly lead to cancer, cardiovascular disease, and HIV.
HAT1 Background
was the first of the HAT enzymes to be identified in yeast [2]. It is lysine specific for newly synthesized histone 4 (H4). One study showed that the deletion of the HAT caused a loss of acetylation on H4K5 and H4K12, leading to the conclusion that HAT1 is the sole enzyme responsible for the evolutionary conserved histone modification.[3] The enzyme is identified as a binding partner for HAT1 to help modulate the substrate specificity of HAT1 [2]. The complex is highly specific for H4K12.
Hat1/Hat2 Complex Structure
Hat1 is not catalytically active until it binds with HAT2 to form the [4]. HAT1 structure, identified as , includes 317 residues and contains the binding site for acetyl-coenzyme A. HAT2 is identified as , which includes 401 residues in a beta-propeller formation with C7 symmetry. Bound to this complex is the histone protein residues 1-38.
The HAT1 and HAT2 interface is stabilized by several interactions of multiple types. Most of these interactions are located in a HAT1 of residues 200-208. This helix is thought to be important for the heterodimer formation as the deletion of the helix abolishes the interaction between HAT1 and HAT2. This suggests that there may be another protein involved, such as the N terminus tail of H4, acting as a linker protein interacting with the complex interface to further stabilize the complex interface [2]. This HAT1/HAT2 interface is stabilized by between the two subunits. There are three major areas where hydrogen bonds are present aids in this complex formation. The side chain atoms of with the main chain nitrogen of Ala202 in HAT1. The side chain of makes hydrogen bonds with Leu288 and Phe205 respectively. The last area of hydrogen bonds between HAT1 and HAT is found between . The at the interface of the complex appears to be critical for the complex formation. This core consists of aromatic amino acids from HAT1 and leucine amino acids from HAT2, however it does not form any obvious ring stacking.
Once the complex has formed, histone 4 and AcetylCoA can begin to interact. The N-terminal segment of H4 that binds with HAT1/HAT2 can be divided into . Similar to 1BOB the N-terminal region of H4 is embedded in a cave between HAT1 and HAT2, though H4 mainly interacts with HAT1. The C-terminal helix of H4 is found inserted into LP2, the N-terminal helix, and C-terminal groove of HAT2. These interactions are strongly stabilized by salt bridge bonds between the histone and the complex. Previous studies suggest that H4K12 inserts into the active site of HAT1 to access AcCoA.[4] The 4PSW structure has H4K12 aligned with the active site and the AcCoA entering the concave groove from the opposite side. This allows the ε-amino group of H4K12 to contact the SH group of CoA.
Acetyl-CoA (CoFactor) Binding Site
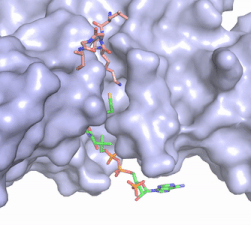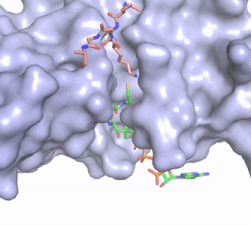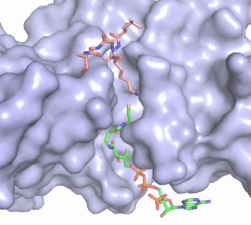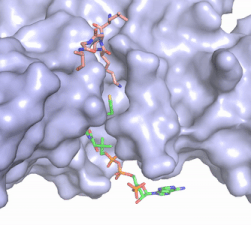
Figure 2. Acetyl-CoA in the binding pocket of HAT1 (Chain A)
The acetyl-CoA HAT1 active site is parallel to the C-terminal domain of the HAT1 protein. Acetyl-CoA fits structurally into the small binding site due to the kinked pantetheine group giving the molecule a bent confirmation. Once bound, most of the acetyl-CoA molecule is buried in the protein, around 60% (Figure 3). Hydrophobic contacts, hydrogen bonds, and salt bridges help to stabilize the protein-ligand interaction.
The β-methyl of the acetyl group interactions in the hydrophobic pocket formed by the side chain of residues: . The carbonyl oxygen of the acetyl group of the main chain Phe-220 and the sulfur of the acetyl-group . In most HAT1 structures, these interactions keep acetyl-CoA in the correct position of the cofactor active site for the transfer of the acetyl-group. In this HAT1 structure, the sulfur acetyl-CoA atom interaction with Asn 258 is unlikely with the distance of 5.5-5.7 angstroms. [5].
Mechanism
After many structural studies, the catalytic mechanism for HAT1 remains unclear. A structural overlay of HAT1 and Gcn5, a better-understood HAT enzyme, found a conserved glutamate residue in the active site of both molecules. Previous studies found that a mutation at the active site glutamate residue greatly alters the catalytic ability of HAT1, proving it to be structurally important. [2] The crystallized structure of the HAT1/HAT2 complex supports, with the proximity of potentially catalytic residues, a mechanism for histone acetylation involving the following residues and cofactor: .
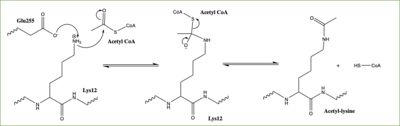
Figure 3: Proposed HAT1 Mechanism. E255 acts as a general base to deprotonate K12 of H4
In this mechanism, the glutamate at residue 255 in the active site of the protein acts as a general base by deprotonating lysine 12 of histone 4 (the numbering of the modified lysine residue on histone 4 is shifted two residues in the featured structure).The deprotonated lysine then acts as a nucleophile and attacks the carbonyl carbon of Acetyl CoA (not shown in the structure), forming a tetrahedral transition state containing an oxyanion. The negative charge on the oxyanion is then shift to down to reform the double bond between the oxygen and carbonyl carbon, breaking the scissle bond between the carbonyl carbon and the sulfur atom of acetyl CoA. The resulting product of this reaction is histone 4 with an acetyl-lysine at residue 12 and coenzyme A.
Inhibition
Although HAT1 was the first histone acetyltransferase enzyme discovered, it is difficult to study and is one of the least understood HAT enzymes. While HAT1 has been linked to many disease states, there is no current known inhibitor of HAT1 that exists. Developing an enzyme inhibitor for HAT1 could allow for therapeutic targets in diseases in which HAT1 has been implicated as well as be used as a tool to better understand the specificity and mechanism in which HAT1 acts to modify histones, in particular histone 4 (H4). HAT1 inhibitors containing the first 20 residues of H4, including the target lysine for modification, and acetyl-CoA and found H4K12CoA to act as a competitive inhibitor to both the peptide substrate as well as acetyl-CoA, potentially laying the foundation for new discovery and better understanding of HAT1 [6]
References
- ↑ Watson, J D, et al. Molecular Biology of the Gene (Seventh Edition). (2014) Boston, MA: Benjamin-Cummings Publishing Company.
- ↑ 2.0 2.1 2.2 2.3 Li Y, Zhang L, Liu T, Chai C, Fang Q, Wu H, Agudelo Garcia PA, Han Z, Zong S, Yu Y, Zhang X, Parthun MR, Chai J, Xu RM, Yang M. Hat2p recognizes the histone H3 tail to specify the acetylation of the newly synthesized H3/H4 heterodimer by the Hat1p/Hat2p complex. Genes Dev. 2014 Jun 1;28(11):1217-27. doi: 10.1101/gad.240531.114. Epub 2014 May , 16. PMID:24835250 doi:http://dx.doi.org/10.1101/gad.240531.114
- ↑ Parthun MR, Widom J, Gottschling DE. The major cytoplasmic histone acetyltransferase in yeast: links to chromatin replication and histone metabolism. Cell. 1996 Oct 4;87(1):85-94. PMID:8858151
- ↑ 4.0 4.1 Wu H, Moshkina N, Min J, Zeng H, Joshua J, Zhou MM, Plotnikov AN. Structural basis for substrate specificity and catalysis of human histone acetyltransferase 1. Proc Natl Acad Sci U S A. 2012 Jun 5;109(23):8925-30. Epub 2012 May 21. PMID:22615379 doi:http://dx.doi.org/10.1073/pnas.1114117109
- ↑ Dutnall RN, Tafrov ST, Sternglanz R, Ramakrishnan V. Structure of the yeast histone acetyltransferase Hat1: insights into substrate specificity and implications for the Gcn5-related N-acetyltransferase superfamily. Cold Spring Harb Symp Quant Biol. 1998;63:501-7. PMID:10384314
- ↑ Ngo L, Brown T, Zheng YG. Bisubstrate inhibitors to target histone acetyltransferase 1. Chem Biol Drug Des. 2019 Jan 14. doi: 10.1111/cbdd.13476. PMID:30637990 doi:http://dx.doi.org/10.1111/cbdd.13476
Student Contributors
- Caitlin Gaich
- Jordan Finch
- Morgan Buckley



