We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Lauryn Padgett/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 9: | Line 9: | ||
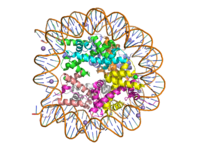
[https://en.wikipedia.org/wiki/Histone Histone proteins] aid in the packing of DNA for the purpose of compacting the genome in the nucleus of the cell and regulating physical accessibility of genes for transcription. The protein itself is an octamer of core proteins H2a, H2b, H3, and H4, which organize into two heterodimers; H1 and H5 act as linker proteins. About 145-157 base pairs wind around a histone heterodimer core. <ref name="DesJarlais">PMID: 26745824</ref> Modifications to histone core proteins can affect the accessibility of transcription factors in the genome, either promoting or inhibiting transcription. Some of these modifications include methylation/demethylation, acetylation/deacetylation, and ubiquitination/deubiquitination. <ref name="Lun">DOI: 10.1016/j.apsb.2013.04.007</ref> | [https://en.wikipedia.org/wiki/Histone Histone proteins] aid in the packing of DNA for the purpose of compacting the genome in the nucleus of the cell and regulating physical accessibility of genes for transcription. The protein itself is an octamer of core proteins H2a, H2b, H3, and H4, which organize into two heterodimers; H1 and H5 act as linker proteins. About 145-157 base pairs wind around a histone heterodimer core. <ref name="DesJarlais">PMID: 26745824</ref> Modifications to histone core proteins can affect the accessibility of transcription factors in the genome, either promoting or inhibiting transcription. Some of these modifications include methylation/demethylation, acetylation/deacetylation, and ubiquitination/deubiquitination. <ref name="Lun">DOI: 10.1016/j.apsb.2013.04.007</ref> | ||
| - | Specifically, histone methylation is associated with gene activation. <ref name="Dong">PMID: 23566087</ref> Many domain families fall under the histone methylase family, one of these enzymes being the <scene name='81/811092/Set7_domain/2'>SET7 domain</scene> family, which can target H3, H4, or H2a. Sites known for gene activation are Lysine-4, Lys-36, and Lys-79 on H3; whereas, methylation at Lys-9 and Lys- 27 on H3 and Lys-20 on H4 are known for gene inactivation.<ref name="Rizzo">PMID: 21847010</ref> Typically, methylation of some of these sites are always present on both active and inactive genes, extra methylations required for activity; specifics of this characteristic depend on site and species of organism. <ref name="Xiao">doi:10.1038/nature01378</ref> Some tumor related genes such as p53 are site specifically methylated to promote biological function <ref name = "Rizzo" />, whereas hypomethylation of CpG is linked to tumor genesis. <ref name="Lun" /> A particular enzyme in the SET7 domain family is lysine methyltransferase, which acts on the histone by adding a methyl group to Lys4 on H3; the addition results in promotion of gene unwinding and gene transcription. <ref name="Xiao" />, <ref name="Dong" /> | + | Specifically, histone methylation is associated with gene activation. <ref name="Dong">PMID: 23566087</ref> Many domain families fall under the histone methylase family, one of these enzymes being the <scene name='81/811092/Set7_domain/2'>SET7 domain</scene> family, which can target H3, H4, or H2a. Sites known for gene activation are Lysine-4, Lys-36, and Lys-79 on H3; whereas, methylation at Lys-9 and Lys- 27 on H3 and Lys-20 on H4 are known for gene inactivation.<ref name="Rizzo">PMID: 21847010</ref> Typically, methylation of some of these sites are always present on both active and inactive genes, extra methylations required for activity; specifics of this characteristic depend on site and species of organism. <ref name="Xiao">doi:10.1038/nature01378</ref> Some tumor related genes such as [https://www.ncbi.nlm.nih.gov/books/NBK22268/ p53] are site specifically methylated to promote biological function <ref name = "Rizzo" />, whereas hypomethylation of [https://en.wikipedia.org/wiki/CpG_site#CpG_islands CpG] is linked to tumor genesis. <ref name="Lun" /> A particular enzyme in the SET7 domain family is lysine methyltransferase, which acts on the histone by adding a methyl group to Lys4 on H3; the addition results in promotion of gene unwinding and gene transcription. <ref name="Xiao" />, <ref name="Dong" /> |
==Lysine Methyltransferase (KMT) Structure== | ==Lysine Methyltransferase (KMT) Structure== | ||
| Line 15: | Line 15: | ||
===Overall Secondary Structure=== | ===Overall Secondary Structure=== | ||
| - | Due to the composition of its [https://en.wikipedia.org/wiki/Protein_secondary_structure secondary structure], KMT is an alpha-beta protein . The helical composition includes 3 <scene name='81/811086/Alpha_helices/4'>alpha helices</scene>, with two residing in the SET domain and one in the C-terminal domain. The alpha helices in the SET domain are two turns while the C-terminal helix is by far the largest with 4 turns. There are also 2 <scene name='81/811086/3-10_helices/4'>3-10 helices</scene> in the SET domain which are each one turn. There are 21 total <scene name='81/811086/Beta_sheets/3'>beta strands</scene> which reside in the N-terminal domain and the SET domain. The beta strands are primarily anti-parallel and multiple antiparallel strands are connected by Type 1 and Type 2 <scene name='81/811086/Beta_turns/3'>beta turns</scene>. | + | Due to the composition of its [https://en.wikipedia.org/wiki/Protein_secondary_structure secondary structure], KMT is an alpha-beta protein <ref name="Xiao" />. The helical composition includes 3 <scene name='81/811086/Alpha_helices/4'>alpha helices</scene>, with two residing in the SET domain and one in the C-terminal domain. The alpha helices in the SET domain are two turns while the C-terminal helix is by far the largest with 4 turns. There are also 2 <scene name='81/811086/3-10_helices/4'>3-10 helices</scene> in the SET domain which are each one turn. There are 21 total <scene name='81/811086/Beta_sheets/3'>beta strands</scene> which reside in the N-terminal domain and the SET domain. The beta strands are primarily anti-parallel and multiple antiparallel strands are connected by Type 1 and Type 2 <scene name='81/811086/Beta_turns/3'>beta turns</scene>. |
===The Active Site=== | ===The Active Site=== | ||
| Line 39: | Line 39: | ||
===Cyproheptadine=== | ===Cyproheptadine=== | ||
| - | Another inhibitor of SET 7/9 is <scene name='81/811086/Cyproheptadine/1'>Cydroheptadine</scene>, a clinically-approved anti-allergy drug that was originally developed as a serotonin and histimine antagonist <ref name="Takemoto">PMID:27088648</ref>. Unlike Sinefungin, it is competitive with the peptide substrates as it binds to the peptide-binding site. The nitrogen of the methylpiperdine ring of cyproheptadine forms a hydrogen bond with Thr286 as well as hydrophobic and van der Waals interactions with the residues surrounding its binding site <ref name=”Takemoto”/>. The binding of cyproheptadine to the catalytic site causes conformational changes of residue Tyr337, an important residue for the formation of the lysine access channel. This movement subsequently causes a conformational change of the beta hairpin, <scene name='81/811086/Altered_beta-hairpin/1'>residues 337-349</scene> | + | Another inhibitor of SET 7/9 is <scene name='81/811086/Cyproheptadine/1'>Cydroheptadine</scene>, a clinically-approved anti-allergy drug that was originally developed as a serotonin and histimine [https://en.wikipedia.org/wiki/Antagonist antagonist] <ref name="Takemoto">PMID:27088648</ref>. Unlike Sinefungin, it is competitive with the peptide substrates as it binds to the peptide-binding site. The nitrogen of the [https://www.koeichem.com/en/product/index.php/item?cell003=Amines&cell004=Piperidine+derivatives&page=8&name=N-Methylpiperidine&id=116&label=1 methylpiperdine] ring of cyproheptadine forms a hydrogen bond with Thr286 as well as hydrophobic and [ https://en.wikipedia.org/wiki/Van_der_Waals_force van der Waals] interactions with the residues surrounding its binding site <ref name=”Takemoto”/ >. The binding of cyproheptadine to the catalytic site causes conformational changes of residue Tyr337, an important residue for the formation of the lysine access channel. This movement subsequently causes a conformational change of the beta hairpin, <scene name='81/811086/Altered_beta-hairpin/1'>residues 337-349</scene> and ultimately generates a large hole adjacent to the lysine access channel, as well as a shift of the C-terminal helix. |
| - | With the revelation of its inhibitory effects on SET7/9, cyproheptadine was used in vitro to treat breast cancer cells ( | + | With the revelation of its inhibitory effects on SET7/9, cyproheptadine was used in vitro to treat breast cancer cells ([https://en.wikipedia.org/wiki/MCF-7 MCF-7] cells). SET 7/9's non-histone activities include the methylation of the estrogen receptor α (ERα), a nuclear receptor and a transcription factor responsible for estrogen-responsive gene regulation. The expression and transcriptional activity of ERα is involved in the carcinogenesis of 70% of breast cancers, making it a major target for hormone therapy. Researchers found that treating the MCF7 cells with cyproheptadine decreased ERα's expression and transcriptional activity which therefore inhibited the estrogen-dependent cell growth. These findings suggest that cyproheptadine could possibly be repurposed to breast cancer therapy in the future. |
==Student Contributors== | ==Student Contributors== | ||
Revision as of 15:18, 26 April 2019
Histone Lysine Methyltransferase: Gene Activator
| |||||||||||
References
- ↑ DesJarlais R, Tummino PJ. Role of Histone-Modifying Enzymes and Their Complexes in Regulation of Chromatin Biology. Biochemistry. 2016 Mar 22;55(11):1584-99. doi: 10.1021/acs.biochem.5b01210. Epub , 2016 Jan 26. PMID:26745824 doi:http://dx.doi.org/10.1021/acs.biochem.5b01210
- ↑ 2.0 2.1 doi: https://dx.doi.org/10.1016/j.apsb.2013.04.007
- ↑ 3.0 3.1 Dong X, Weng Z. The correlation between histone modifications and gene expression. Epigenomics. 2013 Apr;5(2):113-6. doi: 10.2217/epi.13.13. PMID:23566087 doi:http://dx.doi.org/10.2217/epi.13.13
- ↑ 4.0 4.1 Del Rizzo PA, Trievel RC. Substrate and product specificities of SET domain methyltransferases. Epigenetics. 2011 Sep 1;6(9):1059-67. doi: 10.4161/epi.6.9.16069. Epub 2011 Sep, 1. PMID:21847010 doi:http://dx.doi.org/10.4161/epi.6.9.16069
- ↑ 5.0 5.1 5.2 5.3 5.4 5.5 5.6 5.7 Xiao B, Jing C, Wilson JR, Walker PA, Vasisht N, Kelly G, Howell S, Taylor IA, Blackburn GM, Gamblin SJ. Structure and catalytic mechanism of the human histone methyltransferase SET7/9. Nature. 2003 Feb 6;421(6923):652-6. Epub 2003 Jan 22. PMID:12540855 doi:10.1038/nature01378
- ↑ Schluckebier G, Kozak M, Bleimling N, Weinhold E, Saenger W. Differential binding of S-adenosylmethionine S-adenosylhomocysteine and Sinefungin to the adenine-specific DNA methyltransferase M.TaqI. J Mol Biol. 1997 Jan 10;265(1):56-67. PMID:8995524 doi:http://dx.doi.org/10.1006/jmbi.1996.0711
- ↑ Tamura R, Doi S, Nakashima A, Sasaki K, Maeda K, Ueno T, Masaki T. Inhibition of the H3K4 methyltransferase SET7/9 ameliorates peritoneal fibrosis. PLoS One. 2018 May 3;13(5):e0196844. doi: 10.1371/journal.pone.0196844., eCollection 2018. PMID:29723250 doi:http://dx.doi.org/10.1371/journal.pone.0196844
- ↑ Takemoto Y, Ito A, Niwa H, Okamura M, Fujiwara T, Hirano T, Handa N, Umehara T, Sonoda T, Ogawa K, Tariq M, Nishino N, Dan S, Kagechika H, Yamori T, Yokoyama S, Yoshida M. Identification of Cyproheptadine as an Inhibitor of SET Domain Containing Lysine Methyltransferase 7/9 (Set7/9) That Regulates Estrogen-Dependent Transcription. J Med Chem. 2016 Apr 28;59(8):3650-60. doi: 10.1021/acs.jmedchem.5b01732. Epub, 2016 Apr 18. PMID:27088648 doi:http://dx.doi.org/10.1021/acs.jmedchem.5b01732
- ↑ . The binding of cyproheptadine to the catalytic site causes conformational changes of residue Tyr337, an important residue for the formation of the lysine access channel. This movement subsequently causes a conformational change of the beta hairpin, and ultimately generates a large hole adjacent to the lysine access channel, as well as a shift of the C-terminal helix.
With the revelation of its inhibitory effects on SET7/9, cyproheptadine was used in vitro to treat breast cancer cells (MCF-7 cells). SET 7/9's non-histone activities include the methylation of the estrogen receptor α (ERα), a nuclear receptor and a transcription factor responsible for estrogen-responsive gene regulation. The expression and transcriptional activity of ERα is involved in the carcinogenesis of 70% of breast cancers, making it a major target for hormone therapy. Researchers found that treating the MCF7 cells with cyproheptadine decreased ERα's expression and transcriptional activity which therefore inhibited the estrogen-dependent cell growth. These findings suggest that cyproheptadine could possibly be repurposed to breast cancer therapy in the future.
Student Contributors
Lauryn Padgett, Alexandra Pentala, Madeleine Wilson