User:Asif Hossain/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 13: | Line 13: | ||
==HDAC8 Structure== | ==HDAC8 Structure== | ||
| - | The crystal structure of human HDAC8 was determined using x-ray crystallography at a 2.0Å resolution. <ref name="Vannini, A., Volpari, C., Gallinari, P.">Vannini, A., Volpari, C., Gallinari, P., Jones, P., Mattu, M., Carfí, A., ... & Di Marco, S. (2007). Substrate binding to histone deacetylases as shown by the crystal structure of the HDAC8–substrate complex. EMBO reports, 8(9), 879-884. https://doi.org/10.1038/sj.embor.7401047 </ref> The structure includes two structural K ion and one catalytic Zn ion. HDAC8 is bound to a [https://en.wikipedia.org/wiki/P53 p-53] <scene name='81/811084/Ligand/9'>derived diacetylated peptide substrate</scene> as opposed to the natural histone substrate. This peptide includes a <scene name='81/811084/Coumarin/2'>fluorescent coumarin ring</scene> likely used in past kinetic assays. The HDAC8 is made up of a <scene name='81/811084/Beta_sheets/6'>β-sheet</scene> with eight parallel β-strands located between 13 <scene name='81/811084/Alpha_helicesv2/4'>α-helices</scene>. The HDAC8 consists of 377 amino acids. <ref name="Somoza"> Somoza J, Skene R. Structural snapshots of human HDAC8 provide insights into the class I histone deacetylases. Structure, 12(7), 1325-1334.2004. https://doi.org/10.1016/j.str.2004.04.012 </ref> | + | The crystal structure of human HDAC8 was determined using x-ray crystallography at a 2.0Å resolution. <ref name="Vannini, A., Volpari, C., Gallinari, P.">Vannini, A., Volpari, C., Gallinari, P., Jones, P., Mattu, M., Carfí, A., ... & Di Marco, S. (2007). Substrate binding to histone deacetylases as shown by the crystal structure of the HDAC8–substrate complex. EMBO reports, 8(9), 879-884. https://doi.org/10.1038/sj.embor.7401047 </ref> The structure includes two structural K<sup>1+</sup> ion and one catalytic Zn<sup>2+</sup> ion. HDAC8 is bound to a [https://en.wikipedia.org/wiki/P53 p-53] <scene name='81/811084/Ligand/9'>derived diacetylated peptide substrate</scene> as opposed to the natural histone substrate. This peptide includes a <scene name='81/811084/Coumarin/2'>fluorescent coumarin ring</scene> likely used in past kinetic assays. The HDAC8 is made up of a <scene name='81/811084/Beta_sheets/6'>β-sheet</scene> with eight parallel β-strands located between 13 <scene name='81/811084/Alpha_helicesv2/4'>α-helices</scene>. The HDAC8 consists of 377 amino acids. <ref name="Somoza"> Somoza J, Skene R. Structural snapshots of human HDAC8 provide insights into the class I histone deacetylases. Structure, 12(7), 1325-1334.2004. https://doi.org/10.1016/j.str.2004.04.012 </ref> |
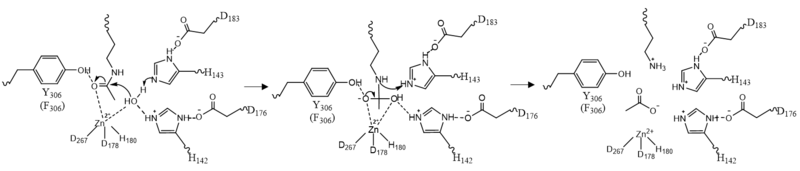
===Zinc Ion=== | ===Zinc Ion=== | ||
Revision as of 17:24, 26 April 2019
Histone Deacetylase 8 (HDAC 8)
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 Vannini, A., Volpari, C., Gallinari, P., Jones, P., Mattu, M., Carfí, A., ... & Di Marco, S. (2007). Substrate binding to histone deacetylases as shown by the crystal structure of the HDAC8–substrate complex. EMBO reports, 8(9), 879-884. https://doi.org/10.1038/sj.embor.7401047
- ↑ DesJarlais, R., & Tummino, P. J. (2016). Role of histone-modifying enzymes and their complexes in regulation of chromatin biology. Biochemistry, 55(11), 1584-1599. https://doi.org/10.1021/acs.biochem.5b01210
- ↑ 3.0 3.1 3.2 3.3 3.4 Somoza J, Skene R. Structural snapshots of human HDAC8 provide insights into the class I histone deacetylases. Structure, 12(7), 1325-1334.2004. https://doi.org/10.1016/j.str.2004.04.012
- ↑ Whitehead, L., Dobler, M. R., Radetich, B., Zhu, Y., Atadja, P. W., Claiborne, T., ... & Shao, W. (2011). Human HDAC isoform selectivity achieved via exploitation of the acetate release channel with structurally unique small molecule inhibitors. Bioorganic & medicinal chemistry, 19(15), 4626-4634. https://doi.org/10.1016/j.bmc.2011.06.030
- ↑ 5.0 5.1 5.2 Vannini, A., Volpari, C., Filocamo, G., Casavola, E. C., Brunetti, M., Renzoni, D., ... & Steinkühler, C. (2004). Crystal structure of a eukaryotic zinc-dependent histone deacetylase, human HDAC8, complexed with a hydroxamic acid inhibitor. Proceedings of the National Academy of Sciences, 101(42), 15064-15069. https://dx.doi.org/10.1073%2Fpnas.0404603101
- ↑ Seto, E., & Yoshida, M. (2014). Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harbor perspectives in biology, 6(4), a018713. https://doi.org/10.1101/cshperspect.a018713
- ↑ 7.0 7.1 Eckschlager T, Plch, J, Stiborova M, Hrabeta J.Histone deacetylase inhibitors as anticancer drugs. International journal of molecular sciences, 18(7), 1414. 2017. https://dx.doi.org/10.3390%2Fijms18071414