User:Caitlin Marie Gaich/Sandbox1
From Proteopedia
(Difference between revisions)
| Line 16: | Line 16: | ||
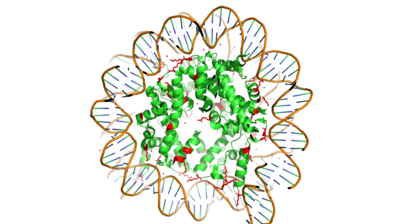
Hat1 is not catalytically active until it binds with HAT2 to form the <scene name='81/811717/Complex/7'>complex</scene> <ref name="Wu">PMID:22615379</ref>. HAT1 structure, identified as <scene name='81/811717/Hat1_-_chain_a/2'>chain A</scene>, includes 317 residues and contains the binding site for acetyl-coenzyme A. HAT2 is identified as <scene name='81/811717/Hat2_-_chain_b/3'>chain B</scene>, which includes 401 residues in a beta-propeller formation with C7 symmetry. Bound to this complex is the histone protein <scene name='81/811717/Histone_4/3'>H4</scene> residues 1-38. | Hat1 is not catalytically active until it binds with HAT2 to form the <scene name='81/811717/Complex/7'>complex</scene> <ref name="Wu">PMID:22615379</ref>. HAT1 structure, identified as <scene name='81/811717/Hat1_-_chain_a/2'>chain A</scene>, includes 317 residues and contains the binding site for acetyl-coenzyme A. HAT2 is identified as <scene name='81/811717/Hat2_-_chain_b/3'>chain B</scene>, which includes 401 residues in a beta-propeller formation with C7 symmetry. Bound to this complex is the histone protein <scene name='81/811717/Histone_4/3'>H4</scene> residues 1-38. | ||
| - | The HAT1 and HAT2 interface is stabilized by several interactions of multiple types. Most of these interactions are located in a HAT1 <scene name='81/811717/Lp1/5'>short, well-ordered helix</scene> of residues 200-208. This helix is thought to be important for the heterodimer formation as the deletion of the helix abolishes the interaction between HAT1 and HAT2. This suggests that there may be another protein involved, such as the N terminus tail of H4, acting as a linker protein interacting with the complex interface to further stabilize the complex interface <ref name="Yang" />. This HAT1/HAT2 interface is stabilized by <scene name='81/811717/Salt_bridges/4'>salt bridges</scene> between the two subunits. There are three major areas where hydrogen bonds are present aids in this complex formation. The side chain atoms of <scene name='81/811717/Tyr199_asp308_ala202/ | + | The HAT1 and HAT2 interface is stabilized by several interactions of multiple types. Most of these interactions are located in a HAT1 <scene name='81/811717/Lp1/5'>short, well-ordered helix</scene> of residues 200-208. This helix is thought to be important for the heterodimer formation as the deletion of the helix abolishes the interaction between HAT1 and HAT2. This suggests that there may be another protein involved, such as the N terminus tail of H4, acting as a linker protein interacting with the complex interface to further stabilize the complex interface <ref name="Yang" />. This HAT1/HAT2 interface is stabilized by <scene name='81/811717/Salt_bridges/4'>salt bridges</scene> between the two subunits. There are three major areas where hydrogen bonds are present aids in this complex formation. The side chain atoms of <scene name='81/811717/Tyr199_asp308_ala202/7'>Tyr199 and Asp308</scene> with the main chain nitrogen of Ala202 in HAT1. The side chain of <scene name='81/811717/Lys211phe205_and_leu288arg282/9'>Lys211 and Arg282</scene> makes hydrogen bonds with Leu288 and Phe205 respectively. The last area of hydrogen bonds between HAT1 and HAT is found between <scene name='81/811717/Serine_hydrogen_bonds/4'>Ser263 and Asp 206</scene>. The <scene name='81/811717/Hydrophobic_core/4'>hydrophobic core</scene> at the interface of the complex appears to be critical for the complex formation. This core consists of aromatic amino acids from HAT1 and leucine amino acids from HAT2, however it does not form any obvious ring stacking. |
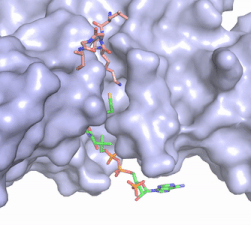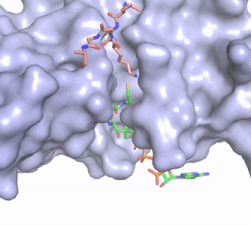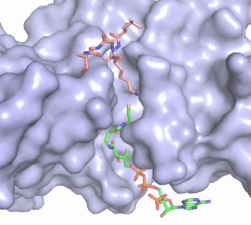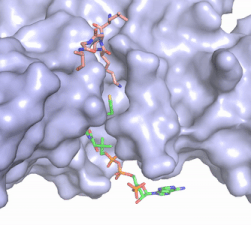
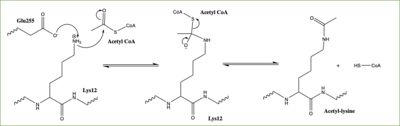
Once the complex has formed, the histone complex can be catalytically active. The N-terminal segment of H4 that binds with HAT1/HAT2 can be divided into <scene name='81/811717/Ntail_regions/3'>three different regions</scene>. Similar to previous HAT1 structures (REFERENCE), the N-terminal region of H4 is buried within a grove on the surface between HAT1 and HAT2, although H4 mostly interacts with HAT1. The C-terminal helix of H4 is found inserted into LP2, the N-terminal helix, and C-terminal groove of HAT2. These interactions are strongly stabilized by salt bridge bonds between the histone and the complex. Previous studies suggest that H4K12 inserts into the active site of HAT1 to access acetyl-CoA. <ref name="Wu"/> | Once the complex has formed, the histone complex can be catalytically active. The N-terminal segment of H4 that binds with HAT1/HAT2 can be divided into <scene name='81/811717/Ntail_regions/3'>three different regions</scene>. Similar to previous HAT1 structures (REFERENCE), the N-terminal region of H4 is buried within a grove on the surface between HAT1 and HAT2, although H4 mostly interacts with HAT1. The C-terminal helix of H4 is found inserted into LP2, the N-terminal helix, and C-terminal groove of HAT2. These interactions are strongly stabilized by salt bridge bonds between the histone and the complex. Previous studies suggest that H4K12 inserts into the active site of HAT1 to access acetyl-CoA. <ref name="Wu"/> | ||
Revision as of 17:25, 26 April 2019
Histone Acetyltransferase HAT1/HAT2 Complex, Saccharomyces cerevisiae
| |||||||||||