We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Madeleine Wilson/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 24: | Line 24: | ||
===The C-Terminal Domain=== | ===The C-Terminal Domain=== | ||
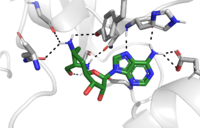
| - | The <scene name='81/811091/C_terminal_domain/1'>C-terminal segment</scene> of lysine methyltransferase is essential for the catalytic activity of the enzyme. Hydrophobic packing of the C-terminal segment (residues 345-366) forms the lysine access channel. Residues 337-349 create a <scene name='81/811086/Beta_hairpin/2'>beta hairpin structure</scene> that stabilizes the orientation of two tyrosine residues Tyr 335 and Tyr337 that form the lysine access channel. The hydrophobic packing of the C-terminal <scene name='81/811091/C_terminal_domain/7'>alpha helix</scene> against beta sheet 19 (specifically residue 299) orient the SAM cofactor so the methyl donating group is oriented toward the lysine access channel. donating group is oriented toward the lysine access channel. | ||
| - | |||
The C-terminal domain of lysine methyltransferase is very important for the catalytic activity of the enzyme. The structures of the <scene name='81/811091/C_terminal_domain/1'>C-terminal domain (residues 345-366)</scene> serve the role of stabilizing the structures in the <scene name='81/811091/C_terminal_domain/16'>SET7 domain (residues 193-344)</scene> in the correct orientation for a reaction in the active site.<ref name="Xiao" /> Hydrophobic interactions in the C-terminal domain are mainly responsible for stabilizing the access channel for the lysine methylation site on histone H3. Residues 337-349 create a <scene name='81/811086/Beta-hairpin/1'>beta-hairpin</scene> that stabilizes the orientation of two tyrosine residues Tyr 335 and Tyr337 that form the lysine access channel. Furthermore, the hydrophobic packing of alpha-helix 3 against beta-sheet 19, specifically <scene name='81/811091/C_terminal_domain/13'>residues Leu357 and Phe 299</scene>, stabilize the orientation of the <scene name='81/811091/C_terminal_domain/14'>SAM cofactor</scene> so that its methyl donating group is oriented toward the lysine access channel. The orientation of the SAM cofactor is further stabilized with its hydrophobic interactions with C-terminal domain residues <scene name='81/811091/C_terminal_domain/18'>Trp352 and Glu356</scene>. <ref name="Xiao" /> | The C-terminal domain of lysine methyltransferase is very important for the catalytic activity of the enzyme. The structures of the <scene name='81/811091/C_terminal_domain/1'>C-terminal domain (residues 345-366)</scene> serve the role of stabilizing the structures in the <scene name='81/811091/C_terminal_domain/16'>SET7 domain (residues 193-344)</scene> in the correct orientation for a reaction in the active site.<ref name="Xiao" /> Hydrophobic interactions in the C-terminal domain are mainly responsible for stabilizing the access channel for the lysine methylation site on histone H3. Residues 337-349 create a <scene name='81/811086/Beta-hairpin/1'>beta-hairpin</scene> that stabilizes the orientation of two tyrosine residues Tyr 335 and Tyr337 that form the lysine access channel. Furthermore, the hydrophobic packing of alpha-helix 3 against beta-sheet 19, specifically <scene name='81/811091/C_terminal_domain/13'>residues Leu357 and Phe 299</scene>, stabilize the orientation of the <scene name='81/811091/C_terminal_domain/14'>SAM cofactor</scene> so that its methyl donating group is oriented toward the lysine access channel. The orientation of the SAM cofactor is further stabilized with its hydrophobic interactions with C-terminal domain residues <scene name='81/811091/C_terminal_domain/18'>Trp352 and Glu356</scene>. <ref name="Xiao" /> | ||
Revision as of 17:34, 26 April 2019
Histone Lysine Methyltransferase: Gene Activator
| |||||||||||
References
- ↑ DesJarlais R, Tummino PJ. Role of Histone-Modifying Enzymes and Their Complexes in Regulation of Chromatin Biology. Biochemistry. 2016 Mar 22;55(11):1584-99. doi: 10.1021/acs.biochem.5b01210. Epub , 2016 Jan 26. PMID:26745824 doi:http://dx.doi.org/10.1021/acs.biochem.5b01210
- ↑ 2.0 2.1 doi: https://dx.doi.org/10.1016/j.apsb.2013.04.007
- ↑ 3.0 3.1 Dong X, Weng Z. The correlation between histone modifications and gene expression. Epigenomics. 2013 Apr;5(2):113-6. doi: 10.2217/epi.13.13. PMID:23566087 doi:http://dx.doi.org/10.2217/epi.13.13
- ↑ 4.0 4.1 Del Rizzo PA, Trievel RC. Substrate and product specificities of SET domain methyltransferases. Epigenetics. 2011 Sep 1;6(9):1059-67. doi: 10.4161/epi.6.9.16069. Epub 2011 Sep, 1. PMID:21847010 doi:http://dx.doi.org/10.4161/epi.6.9.16069
- ↑ 5.0 5.1 5.2 5.3 5.4 5.5 5.6 5.7 Xiao B, Jing C, Wilson JR, Walker PA, Vasisht N, Kelly G, Howell S, Taylor IA, Blackburn GM, Gamblin SJ. Structure and catalytic mechanism of the human histone methyltransferase SET7/9. Nature. 2003 Feb 6;421(6923):652-6. Epub 2003 Jan 22. PMID:12540855 doi:10.1038/nature01378
- ↑ Schluckebier G, Kozak M, Bleimling N, Weinhold E, Saenger W. Differential binding of S-adenosylmethionine S-adenosylhomocysteine and Sinefungin to the adenine-specific DNA methyltransferase M.TaqI. J Mol Biol. 1997 Jan 10;265(1):56-67. PMID:8995524 doi:http://dx.doi.org/10.1006/jmbi.1996.0711
- ↑ Tamura R, Doi S, Nakashima A, Sasaki K, Maeda K, Ueno T, Masaki T. Inhibition of the H3K4 methyltransferase SET7/9 ameliorates peritoneal fibrosis. PLoS One. 2018 May 3;13(5):e0196844. doi: 10.1371/journal.pone.0196844., eCollection 2018. PMID:29723250 doi:http://dx.doi.org/10.1371/journal.pone.0196844
Student Contributors
Lauryn Padgett, Alexandra Pentala, Madeleine Wilson