We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Courtney Brown/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 3: | Line 3: | ||
==Introduction== | ==Introduction== | ||
===Histones=== | ===Histones=== | ||
| - | [https://en.wikipedia.org/wiki/Histone Histones] are basic proteins that interact with nuclear DNA to help condense this DNA into [https://en.wikipedia.org/wiki/Chromatin chromatin]. [https://en.wikipedia.org/wiki/Nucleosome Nucleosomes] are chromatin beads made up of this nuclear DNA wrapped around eight histone proteins, or a [https://en.wikipedia.org/wiki/Histone_octamer histone octamer] in order to fit into the nucleus<ref name="Histones"> Histones | Learn Science at Scitable https://www.nature.com/scitable/definition/histone-histones-57</ref>. The chromatin found inside the nucleus are catagorized into [https://en.wikipedia.org/wiki/Heterochromatin heterochromatin], which are compact and transcriptionally silent, and [https://en.wikipedia.org/wiki/Euchromatin euchromatin], which are less compact and transcriptionally active <ref name="Chromatin"> What is chromatin, heterochromatin and euchromatin? MBInfo https://www.mechanobio.info/genome-regulation/what-is-chromatin-heterochromatin-and-euchromatin</ref>. Modifying histones is a type of [https://en.wikipedia.org/wiki/Epigenetics epigenetics], where changes are made in gene expression without altering the DNA sequence. Four different examples of histone modifications include [https://en.wikipedia.org/wiki/Histone_acetylation_and_deacetylation histone acetylation, histone deacetylation], [https://en.wikipedia.org/wiki/Histone_methylation histone methylation] and [https://en.wikipedia.org/wiki/Demethylase histone demethylation]<ref name="Histones" /> . | + | [https://en.wikipedia.org/wiki/Histone Histones] are basic proteins that interact with nuclear DNA to help condense this DNA into [https://en.wikipedia.org/wiki/Chromatin chromatin]. [https://en.wikipedia.org/wiki/Nucleosome Nucleosomes] are chromatin beads made up of this nuclear DNA wrapped around eight histone proteins, or a [https://en.wikipedia.org/wiki/Histone_octamer histone octamer] in order to fit into the nucleus<ref name="Histones"> Histones | Learn Science at Scitable https://www.nature.com/scitable/definition/histone-histones-57</ref>. The chromatin found inside the nucleus are catagorized into [https://en.wikipedia.org/wiki/Heterochromatin heterochromatin], which are compact and transcriptionally silent, and [https://en.wikipedia.org/wiki/Euchromatin euchromatin], which are less compact and transcriptionally active<ref name="Chromatin"> What is chromatin, heterochromatin and euchromatin? MBInfo https://www.mechanobio.info/genome-regulation/what-is-chromatin-heterochromatin-and-euchromatin</ref>. Modifying histones is a type of [https://en.wikipedia.org/wiki/Epigenetics epigenetics], where changes are made in gene expression without altering the DNA sequence. Four different examples of histone modifications include [https://en.wikipedia.org/wiki/Histone_acetylation_and_deacetylation histone acetylation, histone deacetylation], [https://en.wikipedia.org/wiki/Histone_methylation histone methylation] and [https://en.wikipedia.org/wiki/Demethylase histone demethylation]<ref name="Histones" /> . |
===Histone Deacetylases (HDACs)=== | ===Histone Deacetylases (HDACs)=== | ||
| - | HDACs are versatile enzymes that participate in regulating histone-DNA interactions, along with non-histone proteins, the cell cycle, cell differentiation and DNA-histone interactions. Histone acetylation is the reversal process, performed by [https://en.wikipedia.org/wiki/Histone_acetyltransferase HATs]. HDACs perform ε-amino-lysine deacetylation on acetylated lysines, the amount and residue number of which are unclear, within the histone <ref name="Seto">PMID:24691964</ref>. | + | HDACs are versatile enzymes that participate in regulating histone-DNA interactions, along with non-histone proteins, the cell cycle, cell differentiation and DNA-histone interactions. Histone acetylation is the reversal process, performed by [https://en.wikipedia.org/wiki/Histone_acetyltransferase HATs]. HDACs perform ε-amino-lysine deacetylation on acetylated lysines, the amount and residue number of which are unclear, within the histone<ref name="Seto">PMID:24691964</ref>. |
There are different classes of HDACs based on phylogenetic analysis: Class I (HDACs 1-3 and 8), Class II (HDACs 4-7, 9 and 10), Class III (Sirtuin deacetylases), and Class IV (HDAC 11)<ref name="Vanninni">PMID:17721440</ref>. | There are different classes of HDACs based on phylogenetic analysis: Class I (HDACs 1-3 and 8), Class II (HDACs 4-7, 9 and 10), Class III (Sirtuin deacetylases), and Class IV (HDAC 11)<ref name="Vanninni">PMID:17721440</ref>. | ||
| Line 17: | Line 17: | ||
The structure of HDAC8 was determined via hanging-drop crystallization using a Tyr306Phe mutation (PDB: 2v5w), which was inactive. This allowed the structure to be captured with the substrate bound. The crystallization conditions were: 50 mM Tris–HCl (pH 8.0), 50 mM MgCl<sub>2</sub>, 10% polyethylene glycol 4000, 2 mM tri(2-carboxyethyl)phosphin and 30 mM glycyl-glycyl-glycine at 22°C. The structure was determined at 2.0 angstroms<ref name="Vanninni" />. | The structure of HDAC8 was determined via hanging-drop crystallization using a Tyr306Phe mutation (PDB: 2v5w), which was inactive. This allowed the structure to be captured with the substrate bound. The crystallization conditions were: 50 mM Tris–HCl (pH 8.0), 50 mM MgCl<sub>2</sub>, 10% polyethylene glycol 4000, 2 mM tri(2-carboxyethyl)phosphin and 30 mM glycyl-glycyl-glycine at 22°C. The structure was determined at 2.0 angstroms<ref name="Vanninni" />. | ||
| - | HDAC8 exists as a dimer in the crystal structure, is 388 residues long and consists of one eight-stranded parallel <scene name='81/811716/Beta_sheet/4'>β-sheet</scene> surrounded by 11 <scene name='81/811716/Alpha_helices/2'>α-helices</scene> <ref name="Vanninni" />. HDAC8 is the only functional HDAC that is found to be a single polypeptide instead of being high molecular-weight multi-protein complexes<ref name="Vanninni" />. | + | HDAC8 exists as a dimer in the crystal structure, is 388 residues long and consists of one eight-stranded parallel <scene name='81/811716/Beta_sheet/4'>β-sheet</scene> surrounded by 11 <scene name='81/811716/Alpha_helices/2'>α-helices</scene><ref name="Vanninni" />. HDAC8 is the only functional HDAC that is found to be a single polypeptide instead of being high molecular-weight multi-protein complexes<ref name="Vanninni" />. |
===The Ligand=== | ===The Ligand=== | ||
| Line 27: | Line 27: | ||
===Potassium Ions=== | ===Potassium Ions=== | ||
| - | There are two potassium ions within HDAC8, which increase the catalytic activity and stability of the enzyme overall. The first <scene name='81/811715/Potassium_binding_site/5'>potassium ion</scene> is closest to the active site and interacts with active site residues (side chain oxygen of Ser199/Asp176 and the backbone oxygen of Asp176/Asp178/His180/Leu200), with an octahedral geometry. The second potassium ion is about 20 Å from the catalytic center and regulates the enzymatic activity by an allosteric effect <ref name="Chen">PMID:25060069</ref>. | + | There are two potassium ions within HDAC8, which increase the catalytic activity and stability of the enzyme overall. The first <scene name='81/811715/Potassium_binding_site/5'>potassium ion</scene> is closest to the active site and interacts with active site residues (side chain oxygen of Ser199/Asp176 and the backbone oxygen of Asp176/Asp178/His180/Leu200), with an octahedral geometry. The second potassium ion is about 20 Å from the catalytic center and regulates the enzymatic activity by an allosteric effect<ref name="Chen">PMID:25060069</ref>. |
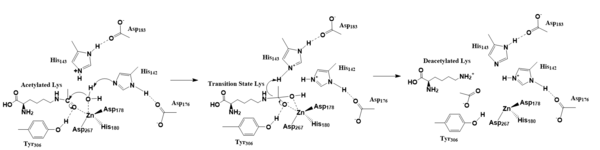
==Active Site== | ==Active Site== | ||
| Line 43: | Line 43: | ||
==Inhibition== | ==Inhibition== | ||
===HDAC Inhibitors=== | ===HDAC Inhibitors=== | ||
| - | HDAC inhibitors ([https://friedreichsataxianews.com/friedreichs-ataxia-experimental-treatments/histone-deacetylase-inhibitors/ HDAC inhibitors]) are a class of compounds that deactivate histone deacetylases. While a variety of these inhibitors are used to target different HDACs, common structural motifs among them include a zinc-binding moiety in the catalytic pocket opposite of the capping group, and a straight chain alkyl, vinyl, or aryl linker connecting the catalytic pocket and capping group, usually a hydrophobic chain of six carbons. HDAC inhibitors bind and deactivate the HDACs through the amino acid sequence of the rim surrounding the catalytic site of the different HDACs (<scene name='81/811716/Hdac8_hydroxamicacid/6'>HDAC8 with Inhibitor</scene>)<ref name="Tabackman">PMID:27374062</ref>. The capping group is generally hydrophobic and interacts with the rim amino acids of the HDAC. Binding occurs via interaction between conserved sections of HDAC active sites and the alkyl, vinyl, or aryl functional groups of the HDAC inhibitors. The zinc binding moiety catalyzes hydrolysis of the acetyl-lysine bond at the bottom of the catalytic pocket, allowing for deactivation of the HDAC <ref name="Marks">PMID:20594930</ref>. (<scene name='81/811716/Hdac8activesiteinhib/5'>Catalytic Pocket Bound by Inhibitor</scene>) <ref name="Vannini">PMID:15477595</ref>. | + | HDAC inhibitors ([https://friedreichsataxianews.com/friedreichs-ataxia-experimental-treatments/histone-deacetylase-inhibitors/ HDAC inhibitors]) are a class of compounds that deactivate histone deacetylases. While a variety of these inhibitors are used to target different HDACs, common structural motifs among them include a zinc-binding moiety in the catalytic pocket opposite of the capping group, and a straight chain alkyl, vinyl, or aryl linker connecting the catalytic pocket and capping group, usually a hydrophobic chain of six carbons. HDAC inhibitors bind and deactivate the HDACs through the amino acid sequence of the rim surrounding the catalytic site of the different HDACs (<scene name='81/811716/Hdac8_hydroxamicacid/6'>HDAC8 with Inhibitor</scene>)<ref name="Tabackman">PMID:27374062</ref>. The capping group is generally hydrophobic and interacts with the rim amino acids of the HDAC. Binding occurs via interaction between conserved sections of HDAC active sites and the alkyl, vinyl, or aryl functional groups of the HDAC inhibitors. The zinc binding moiety catalyzes hydrolysis of the acetyl-lysine bond at the bottom of the catalytic pocket, allowing for deactivation of the HDAC<ref name="Marks">PMID:20594930</ref>. (<scene name='81/811716/Hdac8activesiteinhib/5'>Catalytic Pocket Bound by Inhibitor</scene>)<ref name="Vannini">PMID:15477595</ref>. |
===Clinical Application=== | ===Clinical Application=== | ||
Revision as of 17:39, 26 April 2019
The Human Histone H3/K9 Deacetylase, HDAC8
| |||||||||||
References
- ↑ 1.0 1.1 Histones | Learn Science at Scitable https://www.nature.com/scitable/definition/histone-histones-57
- ↑ What is chromatin, heterochromatin and euchromatin? MBInfo https://www.mechanobio.info/genome-regulation/what-is-chromatin-heterochromatin-and-euchromatin
- ↑ Seto E, Yoshida M. Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harb Perspect Biol. 2014 Apr 1;6(4):a018713. doi:, 10.1101/cshperspect.a018713. PMID:24691964 doi:http://dx.doi.org/10.1101/cshperspect.a018713
- ↑ 4.00 4.01 4.02 4.03 4.04 4.05 4.06 4.07 4.08 4.09 4.10 4.11 4.12 Vannini A, Volpari C, Gallinari P, Jones P, Mattu M, Carfi A, De Francesco R, Steinkuhler C, Di Marco S. Substrate binding to histone deacetylases as shown by the crystal structure of the HDAC8-substrate complex. EMBO Rep. 2007 Sep;8(9):879-84. Epub 2007 Aug 10. PMID:17721440
- ↑ Chen K, Zhang X, Wu YD, Wiest O. Inhibition and mechanism of HDAC8 revisited. J Am Chem Soc. 2014 Aug 20;136(33):11636-43. doi: 10.1021/ja501548p. Epub 2014, Aug 7. PMID:25060069 doi:http://dx.doi.org/10.1021/ja501548p
- ↑ Tabackman AA, Frankson R, Marsan ES, Perry K, Cole KE. Structure of 'linkerless' hydroxamic acid inhibitor-HDAC8 complex confirms the formation of an isoform-specific subpocket. J Struct Biol. 2016 Sep;195(3):373-8. doi: 10.1016/j.jsb.2016.06.023. Epub 2016, Jun 29. PMID:27374062 doi:http://dx.doi.org/10.1016/j.jsb.2016.06.023
- ↑ 7.0 7.1 Marks PA. Histone deacetylase inhibitors: a chemical genetics approach to understanding cellular functions. Biochim Biophys Acta. 2010 Oct-Dec;1799(10-12):717-25. doi:, 10.1016/j.bbagrm.2010.05.008. Epub 2010 Jun 8. PMID:20594930 doi:http://dx.doi.org/10.1016/j.bbagrm.2010.05.008
- ↑ Vannini A, Volpari C, Filocamo G, Casavola EC, Brunetti M, Renzoni D, Chakravarty P, Paolini C, De Francesco R, Gallinari P, Steinkuhler C, Di Marco S. Crystal structure of a eukaryotic zinc-dependent histone deacetylase, human HDAC8, complexed with a hydroxamic acid inhibitor. Proc Natl Acad Sci U S A. 2004 Oct 19;101(42):15064-9. Epub 2004 Oct 11. PMID:15477595
Student Contributors
- Courtney Brown
- Cassandra Marsh
- Carolyn Hurdle