User:Caitlin Marie Gaich/Sandbox1
From Proteopedia
(Difference between revisions)
| Line 7: | Line 7: | ||
=Histone Modification= | =Histone Modification= | ||
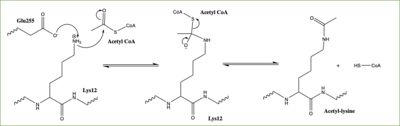
| - | Histones can be modified in a variety of ways, including: methylation, demethylation, acetylation, deacetylation and many others, all leading to either the condensation or relaxation of DNA and as a consequence turning on or off DNA transcription. Histone acetylation is histone modification that involves the transfer of an acetyl group from Acetyl Coenzyme A (acetyl-CoA) to an ε-amino group of a lysine residue on a histone. This reaction is done by various histone acetyltransferase (HAT) enzymes. The specific histone acetylation modification is an important [https://en.wikipedia.org/wiki/Epigenetics epigenetic] marker. It plays a role in RNA synthesis and there a known correlation between gene activity and histone acetylation. Any misregulations of the HAT enzyme can possibly lead to cancer, cardiovascular disease, and HIV <ref name="Ngo"> PMID:30637990 </ref>. | + | Histones can be modified in a variety of ways, including: methylation, demethylation, acetylation, deacetylation and many others, all leading to either the condensation or relaxation of DNA and as a consequence turning on or off DNA transcription. Histone acetylation is histone modification that involves the transfer of an acetyl group from Acetyl Coenzyme A (acetyl-CoA) to an ε-amino group of a lysine residue on a histone. This reaction is done by various histone acetyltransferase (HAT) enzymes. The specific histone acetylation modification is an important [https://en.wikipedia.org/wiki/Epigenetics epigenetic] marker. It plays a role in RNA synthesis and there a known correlation between gene activity and histone acetylation. Any misregulations of the HAT enzyme can possibly lead to cancer, cardiovascular disease, and HIV <ref name="Ngo"> PMID: 30637990 </ref>. |
=HAT1 Background = | =HAT1 Background = | ||
| - | <scene name='81/811717/Hat1_with_accoa/3'>HAT1</scene> was the first of the enzymes to be identified (in yeast) in the HAT family of enzymes <ref name="Yang"> PMID:24835250 </ref>. It is lysine specific for newly synthesized histone 4 (H4). One study showed that the deletion of the HAT caused a loss of acetylation on H4K5 and H4K12, leading to the conclusion that HAT1 is the sole enzyme responsible for the evolutionary conserved histone modification.<ref name="Parthun">PMID:8858151</ref> The <scene name='81/811717/Hat2/1'>HAT2</scene> enzyme is identified as a binding partner for HAT1 to help modulate the substrate specificity of HAT1 <ref name="Yang"/>. The complex is highly specific for H4K12. | + | <scene name='81/811717/Hat1_with_accoa/3'>HAT1</scene> was the first of the enzymes to be identified (in yeast) in the HAT family of enzymes <ref name="Yang"> PMID: 24835250 </ref>. It is lysine specific for newly synthesized histone 4 (H4). One study showed that the deletion of the HAT caused a loss of acetylation on H4K5 and H4K12, leading to the conclusion that HAT1 is the sole enzyme responsible for the evolutionary conserved histone modification.<ref name="Parthun"> PMID: 8858151 </ref> The <scene name='81/811717/Hat2/1'>HAT2</scene> enzyme is identified as a binding partner for HAT1 to help modulate the substrate specificity of HAT1 <ref name="Yang"/>. The complex is highly specific for H4K12. |
= Hat1/Hat2 Complex Structure = | = Hat1/Hat2 Complex Structure = | ||
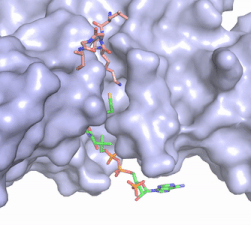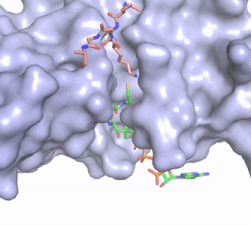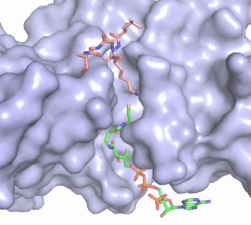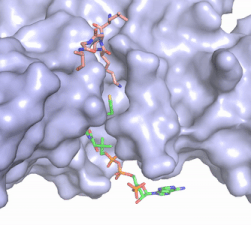
| - | Hat1 is not catalytically active until it binds with HAT2 to form the <scene name='81/811717/Complex/7'>complex</scene> <ref name="Wu">PMID:22615379</ref>. HAT1 structure, identified as <scene name='81/811717/Hat1_-_chain_a/2'>chain A</scene>, includes 317 residues and contains the binding site for acetyl-coenzyme A. HAT2 is identified as <scene name='81/811717/Hat2_-_chain_b/3'>chain B</scene>, which includes 401 residues in a beta-propeller formation with C7 symmetry. Bound to this complex is the histone protein <scene name='81/811717/Histone_4/3'>H4</scene> residues 1-38. | + | Hat1 is not catalytically active until it binds with HAT2 to form the <scene name='81/811717/Complex/7'>complex</scene> <ref name="Wu"> PMID: 22615379 </ref>. HAT1 structure, identified as <scene name='81/811717/Hat1_-_chain_a/2'>chain A</scene>, includes 317 residues and contains the binding site for acetyl-coenzyme A. HAT2 is identified as <scene name='81/811717/Hat2_-_chain_b/3'>chain B</scene>, which includes 401 residues in a beta-propeller formation with C7 symmetry. Bound to this complex is the histone protein <scene name='81/811717/Histone_4/3'>H4</scene> residues 1-38. |
The HAT1 and HAT2 interface is stabilized by several interactions of multiple types. Most of these interactions are located in a HAT1 <scene name='81/811717/Lp1/5'>short, well-ordered helix</scene> of residues 200-208. This helix is thought to be important for the heterodimer formation as the deletion of the helix abolishes the interaction between HAT1 and HAT2. This suggests that there may be another protein involved, such as the N terminus tail of H4, acting as a linker protein interacting with the complex interface to further stabilize the complex interface <ref name="Yang" />. This HAT1/HAT2 interface is stabilized by <scene name='81/811717/Salt_bridges/4'>salt bridges</scene> between the two subunits. There are three major areas where hydrogen bonds are present aids in this complex formation. The side chain atoms of <scene name='81/811717/Tyr199_asp308_ala202/7'>Tyr199 and Asp308</scene> with the main chain nitrogen of Ala202 in HAT1. The side chain of <scene name='81/811717/Lys211phe205_and_leu288arg282/9'>Lys211 and Arg282</scene> makes hydrogen bonds with Leu288 and Phe205 respectively. The last area of hydrogen bonds between HAT1 and HAT is found between <scene name='81/811717/Serine_hydrogen_bonds/4'>Ser263 and Asp 206</scene>. The <scene name='81/811717/Hydrophobic_core/4'>hydrophobic core</scene> at the interface of the complex appears to be critical for the complex formation. This core consists of aromatic amino acids from HAT1 and leucine amino acids from HAT2, however it does not form any obvious ring stacking. | The HAT1 and HAT2 interface is stabilized by several interactions of multiple types. Most of these interactions are located in a HAT1 <scene name='81/811717/Lp1/5'>short, well-ordered helix</scene> of residues 200-208. This helix is thought to be important for the heterodimer formation as the deletion of the helix abolishes the interaction between HAT1 and HAT2. This suggests that there may be another protein involved, such as the N terminus tail of H4, acting as a linker protein interacting with the complex interface to further stabilize the complex interface <ref name="Yang" />. This HAT1/HAT2 interface is stabilized by <scene name='81/811717/Salt_bridges/4'>salt bridges</scene> between the two subunits. There are three major areas where hydrogen bonds are present aids in this complex formation. The side chain atoms of <scene name='81/811717/Tyr199_asp308_ala202/7'>Tyr199 and Asp308</scene> with the main chain nitrogen of Ala202 in HAT1. The side chain of <scene name='81/811717/Lys211phe205_and_leu288arg282/9'>Lys211 and Arg282</scene> makes hydrogen bonds with Leu288 and Phe205 respectively. The last area of hydrogen bonds between HAT1 and HAT is found between <scene name='81/811717/Serine_hydrogen_bonds/4'>Ser263 and Asp 206</scene>. The <scene name='81/811717/Hydrophobic_core/4'>hydrophobic core</scene> at the interface of the complex appears to be critical for the complex formation. This core consists of aromatic amino acids from HAT1 and leucine amino acids from HAT2, however it does not form any obvious ring stacking. | ||
Revision as of 18:30, 26 April 2019
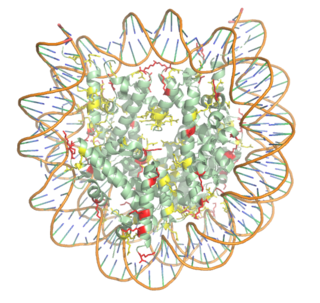
Histone Acetyltransferase HAT1/HAT2 Complex, Saccharomyces cerevisiae
| |||||||||||