User:Carolyn Hurdle/Sandbox 1
From Proteopedia
< User:Carolyn Hurdle(Difference between revisions)
| Line 5: | Line 5: | ||
==Introduction== | ==Introduction== | ||
| - | + | [[Image:Vorinostat_image.png|400 px|right|thumb|Figure 3. The anticancer agent Vorinostat is an example of an HDAC inhibitor that is currently undergoing clinical trial.]] | |
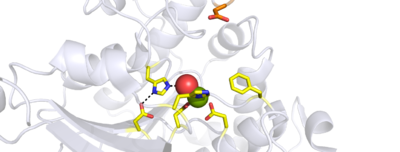
[[Image:Active_site_take_2.png|400 px|right|thumb|Figure 1. View of protein]] | [[Image:Active_site_take_2.png|400 px|right|thumb|Figure 1. View of protein]] | ||
===Histones=== | ===Histones=== | ||
Current revision
The Human Histone H3/K9 Deacetylase, HDAC8
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ Ransey E, Paredes E, Dey SK, Das SR, Heroux A, Macbeth MR. Crystal structure of the Entamoeba histolytica RNA lariat debranching enzyme EhDbr1 reveals a catalytic Zn(2+) /Mn(2+) heterobinucleation. FEBS Lett. 2017 Jul;591(13):2003-2010. doi: 10.1002/1873-3468.12677. Epub 2017, Jun 14. PMID:28504306 doi:http://dx.doi.org/10.1002/1873-3468.12677
- ↑ Vannini A, Volpari C, Gallinari P, Jones P, Mattu M, Carfi A, De Francesco R, Steinkuhler C, Di Marco S. Substrate binding to histone deacetylases as shown by the crystal structure of the HDAC8-substrate complex. EMBO Rep. 2007 Sep;8(9):879-84. Epub 2007 Aug 10. PMID:17721440
Student Contributors
- Carolyn Hurdle
- Courtney Brown
- Cassie Marsh