User:Tereza Čalounová/Sandbox 1
From Proteopedia
m |
|||
| Line 58: | Line 58: | ||
Tafazzin is encoded by a TAZ gene (G4.5). This gene is located on the long arm of the X chromosome at position 28 (Xq28). <ref name="cit6">TAZ gene. In: Www.ncbi.nlm.nih.gov [online]. Rockville Pike: U.S. National Library of Medicine, 2014 [cit. 2019-04-27]. Available at: https://ghr.nlm.nih.gov/gene/TAZ#</ref> | Tafazzin is encoded by a TAZ gene (G4.5). This gene is located on the long arm of the X chromosome at position 28 (Xq28). <ref name="cit6">TAZ gene. In: Www.ncbi.nlm.nih.gov [online]. Rockville Pike: U.S. National Library of Medicine, 2014 [cit. 2019-04-27]. Available at: https://ghr.nlm.nih.gov/gene/TAZ#</ref> | ||
Several isoforms are known. Isoforms that lack the N-terminus are found in leukocytes and fibroblasts, but not in heart and skeletal muscle. <ref name="cit5" /> | Several isoforms are known. Isoforms that lack the N-terminus are found in leukocytes and fibroblasts, but not in heart and skeletal muscle. <ref name="cit5" /> | ||
| - | Human TAZ consists of 11 exons and produces four alternatively spliced mRNA transcripts: a full-length transcript containing all exons (FL), a transcript lacking exon 5 (Δ5), a transcript lacking exon 7 (Δ7) and a transcript lacking both exons 5 and 7 (Δ5Δ7). Of these four alternatively spliced variants of human TAZ, FL and Δ5 encode proteins with transacylase activity, while Δ7 and Δ5Δ7 do not. <ref name="cit8"> | + | Human TAZ consists of 11 exons and produces four alternatively spliced mRNA transcripts: a full-length transcript containing all exons (FL), a transcript lacking exon 5 (Δ5), a transcript lacking exon 7 (Δ7) and a transcript lacking both exons 5 and 7 (Δ5Δ7). Of these four alternatively spliced variants of human TAZ, FL and Δ5 encode proteins with transacylase activity, while Δ7 and Δ5Δ7 do not. <ref name="cit8">PMID: 25941633</ref> |
== Disease - Barth syndrome== | == Disease - Barth syndrome== | ||
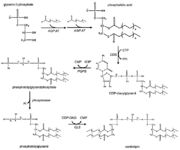
Barth syndrome (BTHS), also known as 3-Methylglutaconic aciduria type II, is an X-linked genetic disorder. The disease is caused by mutation in TAZ gene which encodes for protein tafazzin. <ref name="cit1">https://ghr.nlm.nih.gov/condition/barth-syndrome#</ref> Tafazzin works as an [https://en.wikipedia.org/wiki/Acyltransferase acyltransferase]in complex lipid metabolism, it is responsible for altering immature [https://en.wikipedia.org/wiki/Cardiolipin cardiolipin]- intermediate [https://en.wikipedia.org/wiki/Monolysocardiolipin monolysocardiolipin](with three linoleic acid side chains) (MLCL). <ref name="cit2">https://www.sciencedirect.com/science/article/pii/S092544391830334X?via%3Dihub</ref> <ref name="cit3">https://obgyn.onlinelibrary.wiley.com/doi/full/10.1002/pd.2599</ref> Cardiolipin makes up 20% of mitochondrial lipids and is closely connected with the electron transport chain proteins and the inner membrane structure of the mitochondria. <ref name="cit4">https://www.sciencedirect.com/topics/biochemistry-genetics-and-molecular-biology/tafazzin</ref> Mutations in TAZ gene lead to tafazzin not working properly, immature cardiolipin accumulates whereas the level of cardiolipin is low (mature cardiolopin has four linoleic acid side chains).<ref name="cit2" /><ref name="cit3" /> Mitochondria in affected patients are not having a normal shape and functions. Reduced energy production of mitochondria results in apoptosis of cells in tissues with high energy demands, especially cardiac and skeletal muscles. Moreover abnormally shaped mitochondria in white blood cells may affect their ability to proliferate. This causes [https://en.wikipedia.org/wiki/Neutropenia neutropenia]- decreased amount of white blood cells leading to higher risk of infections. <ref name="cit1" /> | Barth syndrome (BTHS), also known as 3-Methylglutaconic aciduria type II, is an X-linked genetic disorder. The disease is caused by mutation in TAZ gene which encodes for protein tafazzin. <ref name="cit1">https://ghr.nlm.nih.gov/condition/barth-syndrome#</ref> Tafazzin works as an [https://en.wikipedia.org/wiki/Acyltransferase acyltransferase]in complex lipid metabolism, it is responsible for altering immature [https://en.wikipedia.org/wiki/Cardiolipin cardiolipin]- intermediate [https://en.wikipedia.org/wiki/Monolysocardiolipin monolysocardiolipin](with three linoleic acid side chains) (MLCL). <ref name="cit2">https://www.sciencedirect.com/science/article/pii/S092544391830334X?via%3Dihub</ref> <ref name="cit3">https://obgyn.onlinelibrary.wiley.com/doi/full/10.1002/pd.2599</ref> Cardiolipin makes up 20% of mitochondrial lipids and is closely connected with the electron transport chain proteins and the inner membrane structure of the mitochondria. <ref name="cit4">https://www.sciencedirect.com/topics/biochemistry-genetics-and-molecular-biology/tafazzin</ref> Mutations in TAZ gene lead to tafazzin not working properly, immature cardiolipin accumulates whereas the level of cardiolipin is low (mature cardiolopin has four linoleic acid side chains).<ref name="cit2" /><ref name="cit3" /> Mitochondria in affected patients are not having a normal shape and functions. Reduced energy production of mitochondria results in apoptosis of cells in tissues with high energy demands, especially cardiac and skeletal muscles. Moreover abnormally shaped mitochondria in white blood cells may affect their ability to proliferate. This causes [https://en.wikipedia.org/wiki/Neutropenia neutropenia]- decreased amount of white blood cells leading to higher risk of infections. <ref name="cit1" /> | ||
| Line 87: | Line 87: | ||
His 69 – Asp 74 PŘIDAT SCÉNU | His 69 – Asp 74 PŘIDAT SCÉNU | ||
| - | Hijikatu, Yura, Ohara and Go in their work from 2015 predicted the intrinsically unstructured regions present in human tafazzin using 15 available prediction servers, eleven of them consistently predicted that the region encoded by exon 5 is intrinsically unstructured, and no other regions of the tafazzin sequence were consistently predicted as intrinsically unstructured.<ref name=" | + | Hijikatu, Yura, Ohara and Go in their work from 2015 predicted the intrinsically unstructured regions present in human tafazzin using 15 available prediction servers, eleven of them consistently predicted that the region encoded by exon 5 is intrinsically unstructured, and no other regions of the tafazzin sequence were consistently predicted as intrinsically unstructured.<ref name="cit8" /> |
</StructureSection> | </StructureSection> | ||
Revision as of 21:11, 27 April 2019
This is our page where we will share informations about protein Tafazzin as a part of a school project with my classmates. This page is under a construction so please be aware of it. Zde přidám úvod
Tafazzin
| Theoretical Model: The protein structure described on this page was determined theoretically, and hence should be interpreted with caution. |
Tafazzin is a protein located in mitochondrial inner membranes. It is involved in altering cardiolipin. Cardiolipin is key in maintaining mitochondrial shape, energy production, and protein transport within cells. The full-length tafazzin protein contains 292 amino acids and has a molecular weight of 33459 daltons. Mutations in gene associated with this protein can cause Barth Syndrome. Barth syndrome (BTHS),is a genetic disorder diagnosed almost exclusively in males. BTHS is rare, it is estimated to affect 1 in 300,000 to 400,000 individuals worldwide. Males with BTHS have weak heart and skeletal muscles which can lead to heart failure. Another of the symptoms is neutropenia which can lead to infections. [1]
| |||||||||||
References
- ↑ Barth syndrome. In: Ghr.nlm.nih.gov [online]. Rockville Pike: U.S. National Library of Medicine, 2019 [cit. 2019-04-27]. Available at: https://ghr.nlm.nih.gov/condition/barth-syndrome#
- ↑ 2.0 2.1 2.2 2.3 Q16635 (TAZ_HUMAN). In: Https://www.uniprot.org/ [online]. Cambridge, Geneva, Washington: UniProt, 2019 [cit. 2019-04-27]. Available at: https://www.uniprot.org/uniprot/Q16635?fbclid=IwAR3v10lUTRZfb0NFOYKC4wjaherdU9PIVJ8T63jkC9RfNu_5OQ2IpoDR0iY
- ↑ 3.0 3.1 3.2 3.3 CHRISTIE, William. Cardiolipin (Diphosphatidylglycerol). In: Www.lipidhome.co.uk [online]. Hutton: The LipidWeb, 2019 [cit. 2019-04-27]. Available at: http://www.lipidhome.co.uk/lipids/complex/dpg/index.htm
- ↑ 4.0 4.1 4.2 4.3 Houtkooper, R. H., & Vaz, F. M. (2008). Cardiolipin, the heart of mitochondrial metabolism. Cellular and Molecular Life Sciences, 65(16), 2493–2506. doi:10.1007/s00018-008-8030-5
- ↑ Cardiolipin structure. In: Commons.wikimedia.org [online]. San Francisco: Wikimedia, 2019 [cit. 2019-04-27]. Available at: https://commons.wikimedia.org/wiki/File:Cardiolipin_structure.svg?uselang=cs
- ↑ Rochellehx. File:Eukaryotic pathway.jpg. In: Wikimedia Commons [online]. 23. 4. 2009 [cit. 2019-04-27]. Available at: https://commons.wikimedia.org/wiki/File:Eukaryotic_pathway.jpg
- ↑ 7.0 7.1 7.2 THE FUNCTIONS OF CARDIOLIPIN IN CELLULAR METABOLISM – POTENTIAL MODIFIERS OF THE BARTH SYNDROME PHENOTYPE. In: Www.ncbi.nlm.nih.gov [online]. Rockville Pike: Elsevier, 2014 [cit. 2019-04-27]. Available at: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4342993/
- ↑ TAZ gene. In: Www.ncbi.nlm.nih.gov [online]. Rockville Pike: U.S. National Library of Medicine, 2014 [cit. 2019-04-27]. Available at: https://ghr.nlm.nih.gov/gene/TAZ#
- ↑ 9.0 9.1 Hijikata A, Yura K, Ohara O, Go M. Structural and functional analyses of Barth syndrome-causing mutations and alternative splicing in the tafazzin acyltransferase domain. Meta Gene. 2015 Apr 22;4:92-106. doi: 10.1016/j.mgene.2015.04.001. eCollection, 2015 Jun. PMID:25941633 doi:http://dx.doi.org/10.1016/j.mgene.2015.04.001
- ↑ 10.0 10.1 https://ghr.nlm.nih.gov/condition/barth-syndrome#
- ↑ 11.0 11.1 https://www.sciencedirect.com/science/article/pii/S092544391830334X?via%3Dihub
- ↑ 12.0 12.1 https://obgyn.onlinelibrary.wiley.com/doi/full/10.1002/pd.2599
- ↑ https://www.sciencedirect.com/topics/biochemistry-genetics-and-molecular-biology/tafazzin
- ↑ 14.0 14.1 14.2 14.3 https://www.ncbi.nlm.nih.gov/books/NBK247162/
- ↑ 15.0 15.1 https://swissmodel.expasy.org/repository/uniprot/Q16635
- ↑ http://pfam.xfam.org/protein/Q16635
- ↑ https://pfam.xfam.org/family/PF01553#tabview=tab4
- ↑ https://www.ncbi.nlm.nih.gov/pmc/articles/PMC107040/