User:Tereza Čalounová/Sandbox 1
From Proteopedia
m |
m |
||
| Line 79: | Line 79: | ||
== 3D Structure: Homology Model == | == 3D Structure: Homology Model == | ||
| - | No empirical 3D structure for human tafazzin protein (UniProt Q16635) is available in April, 2019. In view of this, homology model was constructed using Swiss-Model server. <ref name="cit17">https://swissmodel.expasy.org/repository/uniprot/Q16635</ref> The closest related protein to tafazzin with a known 3D structure in a public database is in April 2019 plant glycerol 3-phosphate acyltransferase (G3PAT) which has sequence identity of 18,10 %, sequence similarity is 0,29. <ref name="cit17" /> Tafazzin belongs to a protein superfamily - phospholipid acyltransferases (PF01553).<ref name=" | + | No empirical 3D structure for human tafazzin protein (UniProt Q16635) is available in April, 2019. In view of this, homology model was constructed using Swiss-Model server. <ref name="cit17">Q16635 (TAZ_HUMAN) Homo sapiens (Human). In: Swissmodel.expasy.org/ [online]. Lausanne, Basel: swissmodel, 2019 [cit. 2019-04-27]. Dostupné z: https://swissmodel.expasy.org/repository/uniprot/Q16635</ref> The closest related protein to tafazzin with a known 3D structure in a public database is in April 2019 plant glycerol 3-phosphate acyltransferase (G3PAT) which has sequence identity of 18,10 %, sequence similarity is 0,29. <ref name="cit17" /> Tafazzin belongs to a protein superfamily - phospholipid acyltransferases (PF01553).<ref name="cit25">https://pfam.xfam.org/family/PF01553#tabview=tab0</ref> |
| - | Domains found on sequence of human tafazzin contain transmembrane domain (15-34) and acyltransferase domain (41-215). <ref name="cit19">http://pfam.xfam.org/protein/Q16635</ref> | + | Domains found on sequence of human tafazzin contain transmembrane domain (15-34) and acyltransferase domain (41-215). <ref name="cit19">Protein: TAZ_HUMAN (Q16635). In: Pfam.xfam.org [online]. Heidelberg: EMBL [cit. 2019-04-27]. Dostupné z: http://pfam.xfam.org/protein/Q16635</ref> |
[[Image:ConSurfTafazzin.png|thumb|Output from ConSurf analysis of tafazzin showing HX4D motif conservation.]] | [[Image:ConSurfTafazzin.png|thumb|Output from ConSurf analysis of tafazzin showing HX4D motif conservation.]] | ||
[[Image:HMMLogoAcyltransferase.png|thumb|HMM logo of acyltransferase family (PF01553). <ref name="cit18">https://pfam.xfam.org/family/PF01553#tabview=tab4</ref>]] | [[Image:HMMLogoAcyltransferase.png|thumb|HMM logo of acyltransferase family (PF01553). <ref name="cit18">https://pfam.xfam.org/family/PF01553#tabview=tab4</ref>]] | ||
| Line 86: | Line 86: | ||
In the acyltransferase domain there is a HX<sub>4</sub>D motif - <scene name='81/813423/Tafazzinpredictedcartoonhis69/1'>histidine</scene> and <scene name='81/813423/Tafazzinpredictedcartoonasp74/1'>aspartic acid</scene> residues separated by four less conserved residues. Sequence analysis of membrane-bound glycerolipid acyltransferases revealed that proteins from the bacterial, plant, and animal kingdoms share this highly conserved domain containing <scene name='81/813423/Tafazzinpredictedcartoonhx4d/1'>HX<sub>4</sub>D motif</scene>. | In the acyltransferase domain there is a HX<sub>4</sub>D motif - <scene name='81/813423/Tafazzinpredictedcartoonhis69/1'>histidine</scene> and <scene name='81/813423/Tafazzinpredictedcartoonasp74/1'>aspartic acid</scene> residues separated by four less conserved residues. Sequence analysis of membrane-bound glycerolipid acyltransferases revealed that proteins from the bacterial, plant, and animal kingdoms share this highly conserved domain containing <scene name='81/813423/Tafazzinpredictedcartoonhx4d/1'>HX<sub>4</sub>D motif</scene>. | ||
<ref name="cit21">https://www.ncbi.nlm.nih.gov/pmc/articles/PMC107040/</ref> | <ref name="cit21">https://www.ncbi.nlm.nih.gov/pmc/articles/PMC107040/</ref> | ||
| - | His 69 – Asp 74 PŘIDAT SCÉNU | ||
Hijikatu, Yura, Ohara and Go in their work from 2015 predicted the intrinsically unstructured regions present in human tafazzin using 15 available prediction servers, eleven of them consistently predicted that the region encoded by exon 5 is intrinsically unstructured, and no other regions of the tafazzin sequence were consistently predicted as intrinsically unstructured.<ref name="cit8" /> | Hijikatu, Yura, Ohara and Go in their work from 2015 predicted the intrinsically unstructured regions present in human tafazzin using 15 available prediction servers, eleven of them consistently predicted that the region encoded by exon 5 is intrinsically unstructured, and no other regions of the tafazzin sequence were consistently predicted as intrinsically unstructured.<ref name="cit8" /> | ||
Revision as of 21:45, 27 April 2019
This is our page where we will share informations about protein Tafazzin as a part of a school project with my classmates. This page is under a construction so please be aware of it. Zde přidám úvod
Tafazzin
| Theoretical Model: The protein structure described on this page was determined theoretically, and hence should be interpreted with caution. |
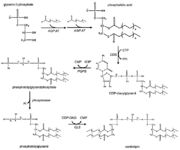
Tafazzin is a protein located in mitochondrial inner membranes. It is involved in altering cardiolipin. Cardiolipin is key in maintaining mitochondrial shape, energy production, and protein transport within cells. The full-length tafazzin protein contains 292 amino acids and has a molecular weight of 33459 daltons. Mutations in gene associated with this protein can cause Barth Syndrome. Barth syndrome (BTHS),is a genetic disorder diagnosed almost exclusively in males. BTHS is rare, it is estimated to affect 1 in 300,000 to 400,000 individuals worldwide. Males with BTHS have weak heart and skeletal muscles which can lead to heart failure. Another of the symptoms is neutropenia which can lead to infections. [1]
| |||||||||||
References
- ↑ 1.0 1.1 1.2 Barth syndrome. In: Ghr.nlm.nih.gov [online]. Rockville Pike: U.S. National Library of Medicine, 2019 [cit. 2019-04-27]. Available at: https://ghr.nlm.nih.gov/condition/barth-syndrome#
- ↑ 2.0 2.1 2.2 2.3 Q16635 (TAZ_HUMAN). In: Https://www.uniprot.org/ [online]. Cambridge, Geneva, Washington: UniProt, 2019 [cit. 2019-04-27]. Available at: https://www.uniprot.org/uniprot/Q16635?fbclid=IwAR3v10lUTRZfb0NFOYKC4wjaherdU9PIVJ8T63jkC9RfNu_5OQ2IpoDR0iY
- ↑ 3.0 3.1 3.2 3.3 CHRISTIE, William. Cardiolipin (Diphosphatidylglycerol). In: Www.lipidhome.co.uk [online]. Hutton: The LipidWeb, 2019 [cit. 2019-04-27]. Available at: http://www.lipidhome.co.uk/lipids/complex/dpg/index.htm
- ↑ 4.0 4.1 4.2 4.3 Houtkooper, R. H., & Vaz, F. M. (2008). Cardiolipin, the heart of mitochondrial metabolism. Cellular and Molecular Life Sciences, 65(16), 2493–2506. doi:10.1007/s00018-008-8030-5
- ↑ Cardiolipin structure. In: Commons.wikimedia.org [online]. San Francisco: Wikimedia, 2019 [cit. 2019-04-27]. Available at: https://commons.wikimedia.org/wiki/File:Cardiolipin_structure.svg?uselang=cs
- ↑ Rochellehx. File:Eukaryotic pathway.jpg. In: Wikimedia Commons [online]. 23. 4. 2009 [cit. 2019-04-27]. Available at: https://commons.wikimedia.org/wiki/File:Eukaryotic_pathway.jpg
- ↑ 7.0 7.1 7.2 Raja V, Greenberg ML. The functions of cardiolipin in cellular metabolism-potential modifiers of the Barth syndrome phenotype. Chem Phys Lipids. 2014 Apr;179:49-56. doi: 10.1016/j.chemphyslip.2013.12.009., Epub 2014 Jan 17. PMID:24445246 doi:http://dx.doi.org/10.1016/j.chemphyslip.2013.12.009
- ↑ TAZ gene. In: Www.ncbi.nlm.nih.gov [online]. Rockville Pike: U.S. National Library of Medicine, 2014 [cit. 2019-04-27]. Available at: https://ghr.nlm.nih.gov/gene/TAZ#
- ↑ 9.0 9.1 Hijikata A, Yura K, Ohara O, Go M. Structural and functional analyses of Barth syndrome-causing mutations and alternative splicing in the tafazzin acyltransferase domain. Meta Gene. 2015 Apr 22;4:92-106. doi: 10.1016/j.mgene.2015.04.001. eCollection, 2015 Jun. PMID:25941633 doi:http://dx.doi.org/10.1016/j.mgene.2015.04.001
- ↑ 10.0 10.1 Barth syndrome cells display widespread remodeling of mitochondrial complexes without affecting metabolic flux distribution. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease [online]. 2018, 1864(11), 3650-3658 [cit. 2019-04-27]. Available at: https://www.sciencedirect.com/science/article/pii/S092544391830334X?via%3Dihub
- ↑ 11.0 11.1 Barth syndrome: an X‐linked cause of fetal cardiomyopathy and stillbirth. Prenatal Diagnosis [online]. 2010, 30(10), 970-976 [cit. 2019-04-27]. Available at: https://obgyn.onlinelibrary.wiley.com/doi/full/10.1002/pd.2599
- ↑ Tafazzin. In: Www.sciencedirect.com [online]. Amsterdam: Elsevier, 2014 [cit. 2019-04-27]. Available at: https://www.sciencedirect.com/topics/biochemistry-genetics-and-molecular-biology/tafazzin
- ↑ 13.0 13.1 13.2 Ferreira C, Thompson R, Vernon H. Barth Syndrome PMID:25299040
- ↑ 14.0 14.1 Q16635 (TAZ_HUMAN) Homo sapiens (Human). In: Swissmodel.expasy.org/ [online]. Lausanne, Basel: swissmodel, 2019 [cit. 2019-04-27]. Dostupné z: https://swissmodel.expasy.org/repository/uniprot/Q16635
- ↑ https://pfam.xfam.org/family/PF01553#tabview=tab0
- ↑ Protein: TAZ_HUMAN (Q16635). In: Pfam.xfam.org [online]. Heidelberg: EMBL [cit. 2019-04-27]. Dostupné z: http://pfam.xfam.org/protein/Q16635
- ↑ https://pfam.xfam.org/family/PF01553#tabview=tab4
- ↑ https://www.ncbi.nlm.nih.gov/pmc/articles/PMC107040/