We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Colicin Immunity Protein
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
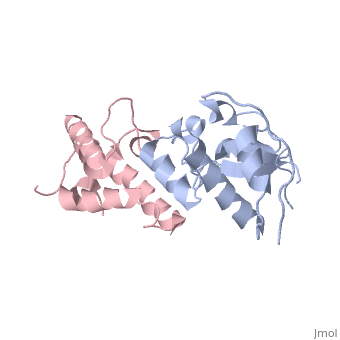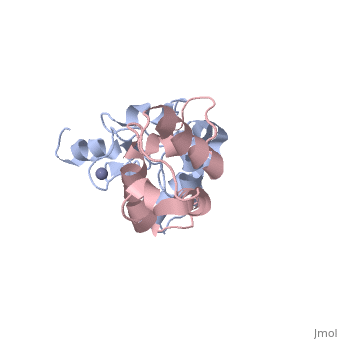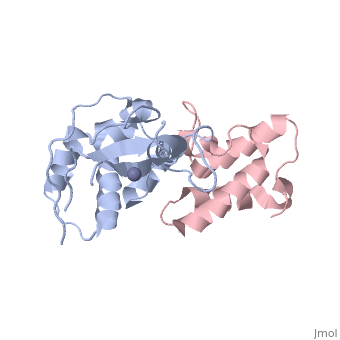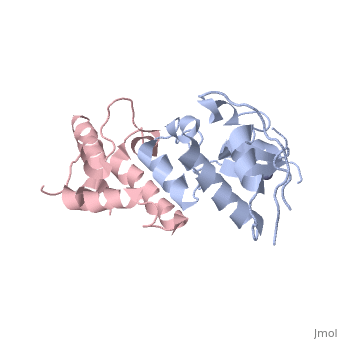
<StructureSection load='3gkl' size='350' scene='' caption='Colicin-E9 immunity protein (grey) complex with colicin-E7 (pink) and Zn+2 ion (grey) (PDB code [[3gkl]])' > | <StructureSection load='3gkl' size='350' scene='' caption='Colicin-E9 immunity protein (grey) complex with colicin-E7 (pink) and Zn+2 ion (grey) (PDB code [[3gkl]])' > | ||
| + | |||
| + | __TOC__ | ||
| + | |||
| + | ==Function== | ||
| + | |||
'''Immunity proteins''' against [[Colicin]]s (EcCIP) are produced by ''E. coli'' alongside the relevant colicin protein (EcCol) to protect the cell from the cytotoxic domain of the colicin. Usually this involves binding to and blocking the active site of the domain, to prevent it from targeting the cells own mechanisms. | '''Immunity proteins''' against [[Colicin]]s (EcCIP) are produced by ''E. coli'' alongside the relevant colicin protein (EcCol) to protect the cell from the cytotoxic domain of the colicin. Usually this involves binding to and blocking the active site of the domain, to prevent it from targeting the cells own mechanisms. | ||
| Line 30: | Line 35: | ||
The <scene name='3gkl/Ali/1'>overall conformation</scene> of the two evolved variants <span style="color:lime;background-color:black;font-weight:bold;">R12-2</span> ([[3gkl]]) and <span style="color:cyan;background-color:black;font-weight:bold;">R12-13</span> ([[3gjn]]) is very similar. The variant <span style="color:lime;background-color:black;font-weight:bold;">R12-2</span> carries <scene name='3gkl/Ali/2'>mutation E41G</scene>. In the <span style="color:yellow;background-color:black;font-weight:bold;">bound wildtype Im9 (yellow)</span> Glu41 makes a <scene name='3gkl/Ali/3'>salt bridge</scene> with the <span style="color:orange;background-color:black;font-weight:bold;">ColE9’s</span> Lys97 ([[1bxi]]). While in the <font color='blueviolet'><b>R12-13</b></font>/<span style="color:cyan;background-color:black;font-weight:bold;">ColE7</span> complex the <font color='blueviolet'><b>closest ColE7 residues</b></font> <scene name='3gkl/Ali/4'>contacting</scene> <span style="color:cyan;background-color:black;font-weight:bold;">R12-13 Glu41</span> are <font color='blueviolet'><b>Thr531 (3.37Å) and Lys528 (8.85Å)</b></font> ([[3gjn]]). In the <span style="color:lime;background-color:black;font-weight:bold;">R12-2</span>/<font color='magenta'><b>ColE7</b></font> complex the <scene name='3gkl/Ali/5'>closest</scene> <font color='magenta'><b>ColE7 residue</b></font> to <span style="color:lime;background-color:black;font-weight:bold;">R12-2 Gly41</span> is <font color='magenta'><b>Thr531 (9.48Å)</b></font> ([[3gkl]]). | The <scene name='3gkl/Ali/1'>overall conformation</scene> of the two evolved variants <span style="color:lime;background-color:black;font-weight:bold;">R12-2</span> ([[3gkl]]) and <span style="color:cyan;background-color:black;font-weight:bold;">R12-13</span> ([[3gjn]]) is very similar. The variant <span style="color:lime;background-color:black;font-weight:bold;">R12-2</span> carries <scene name='3gkl/Ali/2'>mutation E41G</scene>. In the <span style="color:yellow;background-color:black;font-weight:bold;">bound wildtype Im9 (yellow)</span> Glu41 makes a <scene name='3gkl/Ali/3'>salt bridge</scene> with the <span style="color:orange;background-color:black;font-weight:bold;">ColE9’s</span> Lys97 ([[1bxi]]). While in the <font color='blueviolet'><b>R12-13</b></font>/<span style="color:cyan;background-color:black;font-weight:bold;">ColE7</span> complex the <font color='blueviolet'><b>closest ColE7 residues</b></font> <scene name='3gkl/Ali/4'>contacting</scene> <span style="color:cyan;background-color:black;font-weight:bold;">R12-13 Glu41</span> are <font color='blueviolet'><b>Thr531 (3.37Å) and Lys528 (8.85Å)</b></font> ([[3gjn]]). In the <span style="color:lime;background-color:black;font-weight:bold;">R12-2</span>/<font color='magenta'><b>ColE7</b></font> complex the <scene name='3gkl/Ali/5'>closest</scene> <font color='magenta'><b>ColE7 residue</b></font> to <span style="color:lime;background-color:black;font-weight:bold;">R12-2 Gly41</span> is <font color='magenta'><b>Thr531 (9.48Å)</b></font> ([[3gkl]]). | ||
| - | </StructureSection> | ||
==3D structure of Colicin immunity protein== | ==3D structure of Colicin immunity protein== | ||
| + | [[Colicin immunity protein 3D structures]] | ||
| - | + | </StructureSection> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | **[[2xgl]], [[4aeq]] – EcCIPM periplasmic domain<br /> | ||
| - | }} | ||
== References == | == References == | ||
<references/> | <references/> | ||
Revision as of 08:15, 14 May 2019
| |||||||||||
References
- ↑ Meenan NA, Sharma A, Fleishman SJ, Macdonald CJ, Morel B, Boetzel R, Moore GR, Baker D, Kleanthous C. The structural and energetic basis for high selectivity in a high-affinity protein-protein interaction. Proc Natl Acad Sci U S A. 2010 May 17. PMID:20479265
- ↑ Li C, Zhao D, Djebli A, Shoham M. Crystal structure of colicin E3 immunity protein: an inhibitor of a ribosome-inactivating RNase. Structure. 1999 Nov 15;7(11):1365-72. PMID:10574790
- ↑ Yajima S, Inoue S, Ogawa T, Nonaka T, Ohsawa K, Masaki H. Structural basis for sequence-dependent recognition of colicin E5 tRNase by mimicking the mRNA-tRNA interaction. Nucleic Acids Res. 2006;34(21):6074-82. Epub 2006 Nov 11. PMID:17099236 doi:10.1093/nar/gkl729
- ↑ Ko TP, Liao CC, Ku WY, Chak KF, Yuan HS. The crystal structure of the DNase domain of colicin E7 in complex with its inhibitor Im7 protein. Structure. 1999 Jan 15;7(1):91-102. PMID:10368275
- ↑ Montalvao RW, Cavalli A, Salvatella X, Blundell TL, Vendruscolo M. Structure Determination of Protein-Protein Complexes Using NMR Chemical Shifts: Case of an Endonuclease Colicin-Immunity Protein Complex. J Am Chem Soc. 2008 Nov 4. PMID:18980319 doi:10.1021/ja805258z
- ↑ Graille M, Mora L, Buckingham RH, van Tilbeurgh H, de Zamaroczy M. Structural inhibition of the colicin D tRNase by the tRNA-mimicking immunity protein. EMBO J. 2004 Apr 7;23(7):1474-82. Epub 2004 Mar 11. PMID:15014439 doi:10.1038/sj.emboj.7600162
- ↑ Levin KB, Dym O, Albeck S, Magdassi S, Keeble AH, Kleanthous C, Tawfik DS. Following evolutionary paths to protein-protein interactions with high affinity and selectivity. Nat Struct Mol Biol. 2009 Oct;16(10):1049-55. Epub 2009 Sep 13. PMID:19749752 doi:10.1038/nsmb.1670
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Jaime Prilusky, Gemma McGoldrick