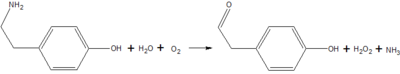
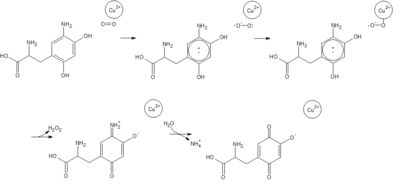
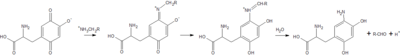Copper Amine Oxidase
From Proteopedia
(Difference between revisions)
| Line 22: | Line 22: | ||
[[Image:2d1w Oxidative Half.png|thumb|center|400px|The oxidative half-reaction, in which topa-quinone is restored to its oxidized form, releasing ammonia and hydrogen peroxide.]] | [[Image:2d1w Oxidative Half.png|thumb|center|400px|The oxidative half-reaction, in which topa-quinone is restored to its oxidized form, releasing ammonia and hydrogen peroxide.]] | ||
<div style="clear:left;"></div> | <div style="clear:left;"></div> | ||
| + | |||
| + | ==3D structures of copper amine oxidase == | ||
| + | [[Copper amine oxidase 3D structures]] | ||
| + | |||
</StructureSection> | </StructureSection> | ||
==3D structures of copper amine oxidase == | ==3D structures of copper amine oxidase == | ||
| Line 30: | Line 34: | ||
*Copper amine oxidase | *Copper amine oxidase | ||
| - | **[[1us1]], [[1pu4]], [[2y73]], [[2y74]] – hCuAO + TPQ | + | **[[6grr]] - hCuAO (mutant) - human<br /> |
| + | **[[1us1]], [[1pu4]], [[2y73]], [[2y74]] – hCuAO + TPQ <br /> | ||
**[[3ala]], [[2c10]] – hCuAO residues 33-763 + TPQ<br /> | **[[3ala]], [[2c10]] – hCuAO residues 33-763 + TPQ<br /> | ||
**[[3loy]], [[2oov]], [[2oqe]] - PaCuAO + TPQ – ''Pichia angusta''<br /> | **[[3loy]], [[2oov]], [[2oqe]] - PaCuAO + TPQ – ''Pichia angusta''<br /> | ||
**[[3n9h]], [[3nbb]], [[3nbj]] – PaCuAO (mutant) + TPQ<br /> | **[[3n9h]], [[3nbb]], [[3nbj]] – PaCuAO (mutant) + TPQ<br /> | ||
**[[2wo0]], [[2wof]], [[2woh]], [[2w0q]], [[1d6u]], [[1spu]], [[1oac]] – EcCuAO residues 31-757 + TPQ – ''Escherichia coli''<br /> | **[[2wo0]], [[2wof]], [[2woh]], [[2w0q]], [[1d6u]], [[1spu]], [[1oac]] – EcCuAO residues 31-757 + TPQ – ''Escherichia coli''<br /> | ||
| + | **[[6ezz]] - EcCuAO (mutant)<br /> | ||
**[[1jrq]], [[1qak]], [[1qal]], [[1qaf]] - EcCuAO (mutant) + TPQ <br /> | **[[1jrq]], [[1qak]], [[1qal]], [[1qaf]] - EcCuAO (mutant) + TPQ <br /> | ||
**[[1w6c]], [[1w6g]], [[1rjo]], [[1av4]], [[1avl]] – AgCuAO + TPQ – ''Arthrobacter globiformis''<br /> | **[[1w6c]], [[1w6g]], [[1rjo]], [[1av4]], [[1avl]] – AgCuAO + TPQ – ''Arthrobacter globiformis''<br /> | ||
Revision as of 09:53, 15 May 2019
| |||||||||||
3D structures of copper amine oxidase
Updated on 15-May-2019
Additional Resources
References
- ↑ 1.0 1.1 Murakawa T, Okajima T, Kuroda S, Nakamoto T, Taki M, Yamamoto Y, Hayashi H, Tanizawa K. Quantum mechanical hydrogen tunneling in bacterial copper amine oxidase reaction. Biochem Biophys Res Commun. 2006 Apr 7;342(2):414-23. Epub 2006 Feb 8. PMID:16487484 doi:10.1016/j.bbrc.2006.01.150
- ↑ Parsons MR, Convery MA, Wilmot CM, Yadav KD, Blakeley V, Corner AS, Phillips SE, McPherson MJ, Knowles PF. Crystal structure of a quinoenzyme: copper amine oxidase of Escherichia coli at 2 A resolution. Structure. 1995 Nov 15;3(11):1171-84. PMID:8591028
- ↑ Mure M, Mills SA, Klinman JP. Catalytic mechanism of the topa quinone containing copper amine oxidases. Biochemistry. 2002 Jul 30;41(30):9269-78. PMID:12135347
- ↑ 4.0 4.1 Grant KL, Klinman JP. Evidence that both protium and deuterium undergo significant tunneling in the reaction catalyzed by bovine serum amine oxidase. Biochemistry. 1989 Aug 8;28(16):6597-605. PMID:2790014
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Raymond Lyle, Alexander Berchansky, OCA, Jaime Prilusky