We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Cystathionine β-synthase
From Proteopedia
(Difference between revisions)
| Line 47: | Line 47: | ||
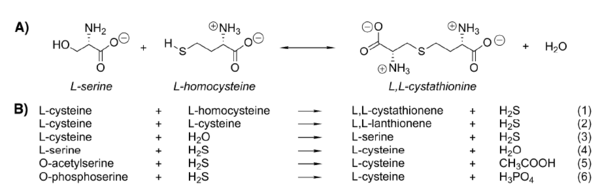
There are more than 100 reported mutations in CBS gene from patients with homocystinuria most of which are missense. Among most frequent mutations is I278T (c.833T>C) in C-terminal domain found in about 25 % of all homocystinuric alleles, and G307S (c.919G>A) located in active site. | There are more than 100 reported mutations in CBS gene from patients with homocystinuria most of which are missense. Among most frequent mutations is I278T (c.833T>C) in C-terminal domain found in about 25 % of all homocystinuric alleles, and G307S (c.919G>A) located in active site. | ||
| - | ''Mutations in active site'' | + | '''Mutations in active site''' |
The active site is accessible only via a narrow channel. Four of the six known point mutations in the active site involve glycine residues: G148R, G305R, G307S and G259S. The residue G259 separates the active site from the heme-binding pocket. The residue G307 lines the entry to the active site cleft and the orientation of G307 do not allow accommodating of the side chain of a serine residue which causes incorporation of the side chain and conformation change in the loop. As the second substrate homocysteine probably binds here the mutation could inhibit binding of homocysteine. | The active site is accessible only via a narrow channel. Four of the six known point mutations in the active site involve glycine residues: G148R, G305R, G307S and G259S. The residue G259 separates the active site from the heme-binding pocket. The residue G307 lines the entry to the active site cleft and the orientation of G307 do not allow accommodating of the side chain of a serine residue which causes incorporation of the side chain and conformation change in the loop. As the second substrate homocysteine probably binds here the mutation could inhibit binding of homocysteine. | ||
| - | ''Mutations in the heme-binding site'' | + | '''Mutations in the heme-binding site''' |
Mutations in this part of the enzyme reduces the ability to bind heme and affects proper folding of CBS. | Mutations in this part of the enzyme reduces the ability to bind heme and affects proper folding of CBS. | ||
| - | ''Mutations in the dimer interface'' | + | '''Mutations in the dimer interface''' |
Numerous CBS mutations (e.g. A114V and G116R) are located at interface of the two monomers and destabilise monomer-monomer interactions and their communication. | Numerous CBS mutations (e.g. A114V and G116R) are located at interface of the two monomers and destabilise monomer-monomer interactions and their communication. | ||
| - | ''Other mutations'' | + | '''Other mutations''' |
Residues in regulatory domain (414-551) bind the allosteric activator S-adenosyl-L-methionine and are responsible for the tetramerization. Mutation R336/H belong to this group of mutations – the side chain of this arginine is packed against the protein surface and the guanidium groups forms a salt bridge to the carboxyl group of D388, residue that does not contribute to any interaction between the two monomers. | Residues in regulatory domain (414-551) bind the allosteric activator S-adenosyl-L-methionine and are responsible for the tetramerization. Mutation R336/H belong to this group of mutations – the side chain of this arginine is packed against the protein surface and the guanidium groups forms a salt bridge to the carboxyl group of D388, residue that does not contribute to any interaction between the two monomers. | ||
The most common mutation, I278T, is located in β-sheet of the C-terminal domain. | The most common mutation, I278T, is located in β-sheet of the C-terminal domain. | ||
Revision as of 21:19, 22 May 2019
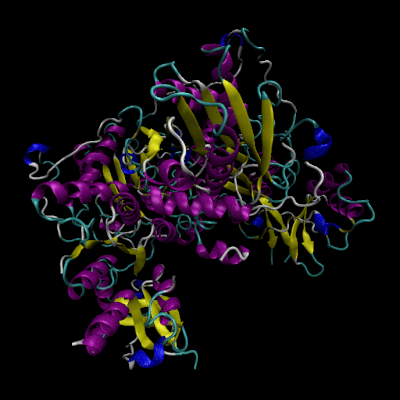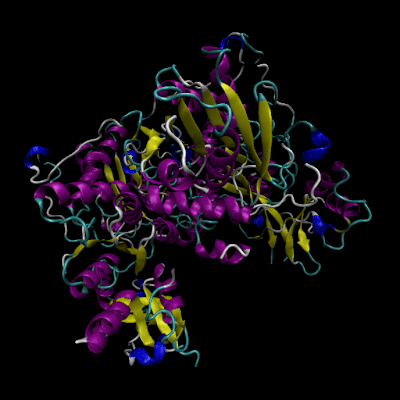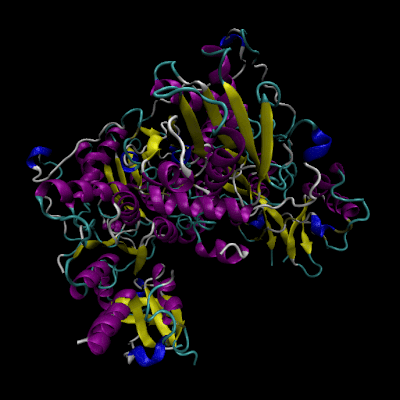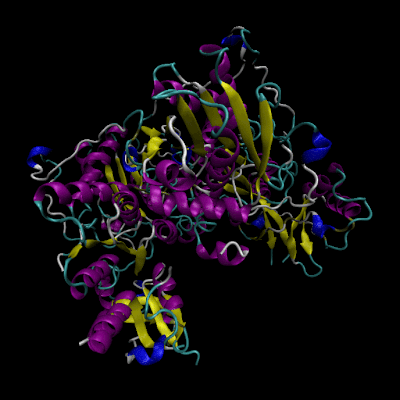
3D Structure of Human Cystathionine β-synthase (4coo)
| |||||||||||
Credits
Article created as an Structural biology of the cell assignment at the Faculty of Science, Charles University, Prague, Czech Republic.
Assignment authors: Jana Křivková, Zdeňka Mauerová, Jan Hamalčík
References
- ↑ Meier M, Janosik M, Kery V, Kraus JP, Burkhard P. Structure of human cystathionine beta-synthase: a unique pyridoxal 5'-phosphate-dependent heme protein. EMBO J. 2001 Aug 1;20(15):3910-6. PMID:11483494 doi:http://dx.doi.org/10.1093/emboj/20.15.3910
- ↑ Meier M, Janosik M, Kery V, Kraus JP, Burkhard P. Structure of human cystathionine beta-synthase: a unique pyridoxal 5'-phosphate-dependent heme protein. EMBO J. 2001 Aug 1;20(15):3910-6. PMID:11483494 doi:http://dx.doi.org/10.1093/emboj/20.15.3910
- ↑ Meier M, Oliveriusova J, Kraus JP, Burkhard P. Structural insights into mutations of cystathionine beta-synthase. Biochim Biophys Acta. 2003 Apr 11;1647(1-2):206-13. PMID:12686134
- ↑ Tu Y, Kreinbring CA, Hill M, Liu C, Petsko GA, McCune CD, Berkowitz DB, Liu D, Ringe D. Crystal Structures of Cystathionine beta-Synthase from Saccharomyces cerevisiae: One Enzymatic Step at a Time. Biochemistry. 2018 Apr 13. doi: 10.1021/acs.biochem.8b00092. PMID:29630349 doi:http://dx.doi.org/10.1021/acs.biochem.8b00092
- ↑ Meier M, Janosik M, Kery V, Kraus JP, Burkhard P. Structure of human cystathionine beta-synthase: a unique pyridoxal 5'-phosphate-dependent heme protein. EMBO J. 2001 Aug 1;20(15):3910-6. PMID:11483494 doi:http://dx.doi.org/10.1093/emboj/20.15.3910
- ↑ Meier M, Janosik M, Kery V, Kraus JP, Burkhard P. Structure of human cystathionine beta-synthase: a unique pyridoxal 5'-phosphate-dependent heme protein. EMBO J. 2001 Aug 1;20(15):3910-6. PMID:11483494 doi:http://dx.doi.org/10.1093/emboj/20.15.3910
- ↑ Meier M, Janosik M, Kery V, Kraus JP, Burkhard P. Structure of human cystathionine beta-synthase: a unique pyridoxal 5'-phosphate-dependent heme protein. EMBO J. 2001 Aug 1;20(15):3910-6. PMID:11483494 doi:http://dx.doi.org/10.1093/emboj/20.15.3910
- ↑ Meier M, Janosik M, Kery V, Kraus JP, Burkhard P. Structure of human cystathionine beta-synthase: a unique pyridoxal 5'-phosphate-dependent heme protein. EMBO J. 2001 Aug 1;20(15):3910-6. PMID:11483494 doi:http://dx.doi.org/10.1093/emboj/20.15.3910
- ↑ Tu Y, Kreinbring CA, Hill M, Liu C, Petsko GA, McCune CD, Berkowitz DB, Liu D, Ringe D. Crystal Structures of Cystathionine beta-Synthase from Saccharomyces cerevisiae: One Enzymatic Step at a Time. Biochemistry. 2018 Apr 13. doi: 10.1021/acs.biochem.8b00092. PMID:29630349 doi:http://dx.doi.org/10.1021/acs.biochem.8b00092
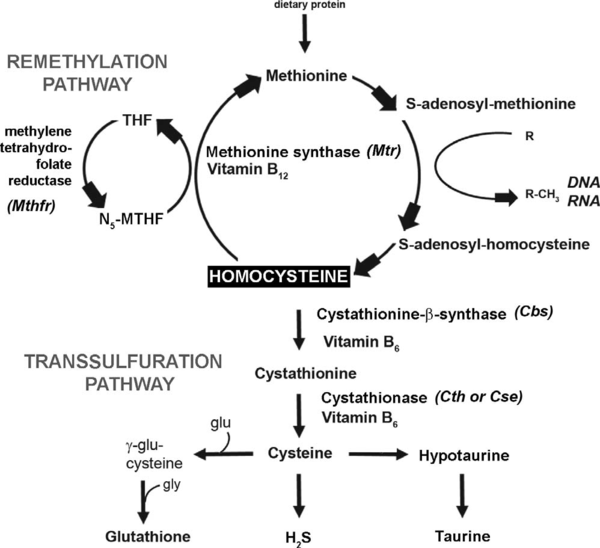
- ↑ Jhee KH, Kruger WD. The role of cystathionine beta-synthase in homocysteine metabolism. Antioxid Redox Signal. 2005 May-Jun;7(5-6):813-22. doi: 10.1089/ars.2005.7.813. PMID:15890029 doi:http://dx.doi.org/10.1089/ars.2005.7.813
- ↑ Ansari R, Mahta A, Mallack E, Luo JJ. Hyperhomocysteinemia and neurologic disorders: a review. J Clin Neurol. 2014 Oct;10(4):281-8. doi: 10.3988/jcn.2014.10.4.281. Epub 2014, Oct 6. PMID:25324876 doi:http://dx.doi.org/10.3988/jcn.2014.10.4.281
- ↑ Miles EW, Kraus JP. Cystathionine beta-synthase: structure, function, regulation, and location of homocystinuria-causing mutations. J Biol Chem. 2004 Jul 16;279(29):29871-4. Epub 2004 Apr 15. PMID:15087459 doi:http://dx.doi.org/10.1074/jbc.R400005200
- ↑ Meier M, Oliveriusova J, Kraus JP, Burkhard P. Structural insights into mutations of cystathionine beta-synthase. Biochim Biophys Acta. 2003 Apr 11;1647(1-2):206-13. PMID:12686134
- ↑ Jhee KH, Kruger WD. The role of cystathionine beta-synthase in homocysteine metabolism. Antioxid Redox Signal. 2005 May-Jun;7(5-6):813-22. doi: 10.1089/ars.2005.7.813. PMID:15890029 doi:http://dx.doi.org/10.1089/ars.2005.7.813